3HJY
 
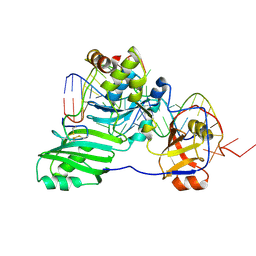 | | Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA | | Descriptor: | 5'-R(*GP*GP*AP*GP*CP*GP*UP*GP*CP*GP*GP*UP*UP*U)-3', 5'-R(*GP*GP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*CP*CP*GP*CP*GP*GP*CP*GP*C)-3', RNA (25-MER), ... | | Authors: | Liang, B, Zhou, J, Kahen, E, Terns, R.M, Terns, M.P, Li, H. | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA
Nat.Struct.Mol.Biol., 16, 2009
|
|
4MZF
 
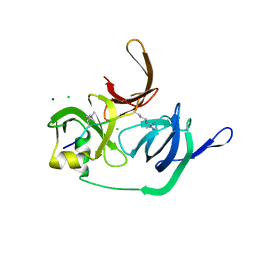 | | Crystal structure of human Spindlin1 bound to histone H3(K4me3-R8me2a) peptide | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Peptide from Histone H3.2, ... | | Authors: | Su, X, Ding, X, Li, H. | | Deposit date: | 2013-09-30 | | Release date: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Molecular basis underlying histone H3 lysine-arginine methylation pattern readout by Spin/Ssty repeats of Spindlin1
Genes Dev., 28, 2014
|
|
4MZH
 
 | |
3NLT
 
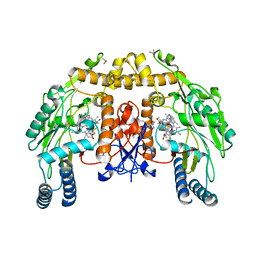 | | Structure of endothelial nitric oxide synthase heme domain complexed with N1-{(3'S,4'S)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}- N2-(3'-fluorophenethyl)ethane-1,2-diamine tetrahydrochloride | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-({(3S,4S)-4-[2-({2,2-difluoro-2-[(2R)-piperidin-2-yl]ethyl}amino)ethoxy]pyrrolidin-3-yl}methyl)-4-methylpyridin-2-amine, CACODYLIC ACID, ... | | Authors: | Xue, F, Li, H, Fang, J, Delker, S.L, Poulos, T.L, Silverman, R.B. | | Deposit date: | 2010-06-21 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Potent, highly selective, and orally bioavailable gem-difluorinated monocationic inhibitors of neuronal nitric oxide synthase.
J.Am.Chem.Soc., 132, 2010
|
|
4N03
 
 | | Fatty acid ABC transporter substrate-binding protein from Thermomonospora curvata | | Descriptor: | 1,2-ETHANEDIOL, ABC-type branched-chain amino acid transport systems periplasmic component-like protein, PALMITIC ACID | | Authors: | Osipiuk, J, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fatty acid ABC transporter substrate-binding protein from Thermomonospora curvata
To be Published
|
|
3NLU
 
 | | Structure of endothelial nitric oxide synthase heme domain complexed with N1-{(3'R,4'R)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine tetrahydrochloride | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[2,2-difluoro-2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Xue, F, Li, H, Fang, J, Delker, S.L, Poulos, T.L, Silverman, R.B. | | Deposit date: | 2010-06-21 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Potent, highly selective, and orally bioavailable gem-difluorinated monocationic inhibitors of neuronal nitric oxide synthase.
J.Am.Chem.Soc., 132, 2010
|
|
3B79
 
 | |
3NKZ
 
 | | The crystal structure of a flagella protein from Yersinia enterocolitica subsp. enterocolitica 8081 | | Descriptor: | Flagellar protein fliT, SULFATE ION, TETRAETHYLENE GLYCOL | | Authors: | Tan, K, Li, H, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | The crystal structure of a flagella protein from Yersinia enterocolitica subsp. enterocolitica 8081
To be Published
|
|
3FH0
 
 | | Crystal structure of putative universal stress protein KPN_01444 - ATPase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, putative universal stress protein KPN_01444 | | Authors: | Chang, C, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-08 | | Release date: | 2008-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of putative universal stress protein KPN_01444 - ATPase
To be Published
|
|
2AG8
 
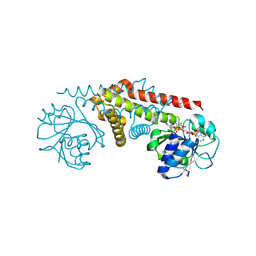 | | NADP complex of Pyrroline-5-carboxylate reductase from Neisseria meningitidis | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, pyrroline-5-carboxylate reductase | | Authors: | Chang, C, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Delta(1)-Pyrroline-5-carboxylate Reductase from Human Pathogens Neisseria meningitides and Streptococcus pyogenes.
J.Mol.Biol., 354, 2005
|
|
3BHG
 
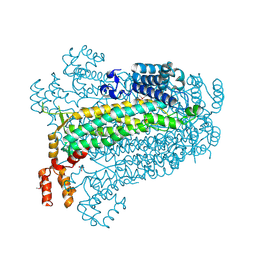 | | Crystal structure of adenylosuccinate lyase from Legionella pneumophila | | Descriptor: | Adenylosuccinate lyase, GLYCEROL, SULFATE ION | | Authors: | Chang, C, Li, H, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-28 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of adenylosuccinate lyase from Legionella pneumophila.
To be Published
|
|
4QYM
 
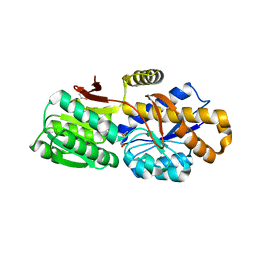 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with methionine | | Descriptor: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, MAGNESIUM ION, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-24 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with methionine
To be Published
|
|
3IF0
 
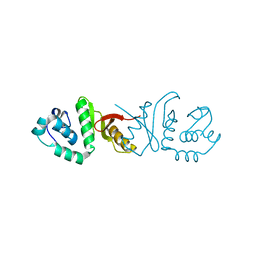 | |
3B48
 
 | |
2FI0
 
 | | The crystal structure of the conserved domain protein from Streptococcus pneumoniae TIGR4 | | Descriptor: | conserved domain protein | | Authors: | Zhang, R, Li, H, Abdullah, J, Collart, F, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-27 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the conserved domain protein from Streptococcus pneumoniae TIGR4
To be Published
|
|
2HKX
 
 | | Structure of CooA mutant (N127L/S128L) from Carboxydothermus hydrogenoformans | | Descriptor: | CARBON MONOXIDE, Carbon monoxide oxidation system transcription regulator CooA-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lanz, N.D, Borjigin, M, Li, H, Kerby, R.L, Poulos, T.L, Roberts, G.P. | | Deposit date: | 2006-07-05 | | Release date: | 2007-03-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based hypothesis on the activation of the CO-sensing transcription factor CooA.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2I0M
 
 | | Crystal structure of the phosphate transport system regulatory protein PhoU from Streptococcus pneumoniae | | Descriptor: | Phosphate transport system protein phoU, ZINC ION | | Authors: | Zhang, R, Li, H, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-10 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the phosphate transport system regulatory protein PhoU from Streptococcus pneumoniae
To be Published, 2006
|
|
2FKZ
 
 | | Reduced (All Ferrous) form of the Azotobacter vinelandii bacterioferritin | | Descriptor: | Bacterioferritin, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Swartz, L, Kunchinskas, M, Li, H, Poulos, T.L, Lanzilotta, W.N. | | Deposit date: | 2006-01-05 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redox-Dependent Structural Changes in the Azotobacter vinelandii Bacterioferritin: New Insights into the Ferroxidase and Iron Transport Mechanism(,).
Biochemistry, 45, 2006
|
|
2HVC
 
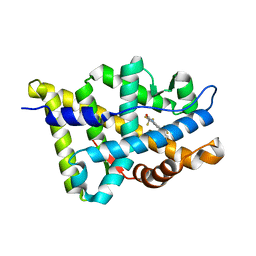 | | The Crystal Structure of Ligand-binding Domain (LBD) of human Androgen Receptor in Complex with a selective modulator LGD2226 | | Descriptor: | 6-[BIS(2,2,2-TRIFLUOROETHYL)AMINO]-4-(TRIFLUOROMETHYL)QUINOLIN-2(1H)-ONE, Androgen receptor | | Authors: | Wang, F, Liu, X.-Q, Li, H, Liang, K.-N, Miner, J.N, Hong, M, Kallel, E.A, van Oeveren, A, Zhi, L, Jiang, T. | | Deposit date: | 2006-07-28 | | Release date: | 2007-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the ligand-binding domain (LBD) of human androgen receptor in complex with a selective modulator LGD2226
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
2I6X
 
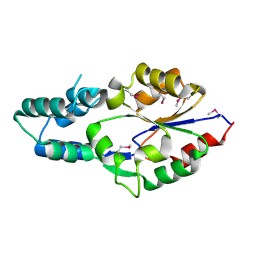 | | The structure of a predicted HAD-like family hydrolase from Porphyromonas gingivalis. | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Cuff, M.E, Mussar, K.E, Li, H, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-29 | | Release date: | 2006-10-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of a predicted HAD-like family hydrolase from Porphyromonas gingivalis. (CASP Target)
To be Published
|
|
2I6D
 
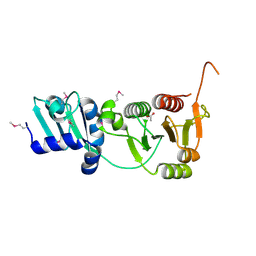 | | The structure of a putative RNA methyltransferase of the TrmH family from Porphyromonas gingivalis. | | Descriptor: | ACETIC ACID, RNA methyltransferase, TrmH family | | Authors: | Cuff, M.E, Mussar, K.E, Li, H, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-28 | | Release date: | 2006-10-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of a putative RNA methyltransferase of the TrmH family from Porphyromonas gingivalis.
TO BE PUBLISHED
|
|
2I9D
 
 | |
2I7H
 
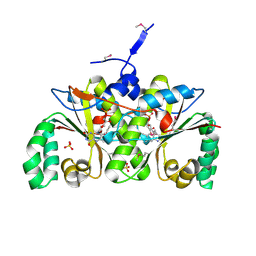 | | Crystal Structure of the Nitroreductase-like Family Protein from Bacillus cereus | | Descriptor: | FLAVIN MONONUCLEOTIDE, Nitroreductase-like family protein, SULFATE ION | | Authors: | Kim, Y, Li, H, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Nitroreductase-like Family Protein from Bacillus cereus
To be Published
|
|
2IS5
 
 | |
2IKB
 
 | |
