1EWS
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE RABBIT KIDNEY DEFENSIN, RK-1 | | Descriptor: | RK-1 DEFENSIN | | Authors: | McManus, A.M, Dawson, N.F, Wade, J.D, Craik, D.J. | | Deposit date: | 2000-04-26 | | Release date: | 2001-05-02 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of RK-1: a novel alpha-defensin peptide.
Biochemistry, 39, 2000
|
|
1AZY
 
 | | STRUCTURAL AND THEORETICAL STUDIES SUGGEST DOMAIN MOVEMENT PRODUCES AN ACTIVE CONFORMATION OF THYMIDINE PHOSPHORYLASE | | Descriptor: | THYMIDINE PHOSPHORYLASE | | Authors: | Pugmire, M.J, Cook, W.J, Jasanoff, A, Walter, M.R, Ealick, S.E. | | Deposit date: | 1997-11-24 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and theoretical studies suggest domain movement produces an active conformation of thymidine phosphorylase.
J.Mol.Biol., 281, 1998
|
|
1A0Q
 
 | | 29G11 COMPLEXED WITH PHENYL [1-(1-N-SUCCINYLAMINO)PENTYL] PHOSPHONATE | | Descriptor: | 29G11 FAB (HEAVY CHAIN), 29G11 FAB (LIGHT CHAIN), PHENYL[1-(N-SUCCINYLAMINO)PENTYL]PHOSPHONATE, ... | | Authors: | Buchbinder, J.L, Stephenson, R.C, Scanlan, T.S, Fletterick, R.J. | | Deposit date: | 1997-12-05 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A comparison of the crystallographic structures of two catalytic antibodies with esterase activity.
J.Mol.Biol., 282, 1998
|
|
1AZN
 
 | | CRYSTAL STRUCTURE OF THE AZURIN MUTANT PHE114ALA FROM PSEUDOMONAS AERUGINOSA AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Tsai, L.-C, Sjolin, L, Langer, V, Pascher, T, Nar, H. | | Deposit date: | 1994-05-27 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the azurin mutant Phe114Ala from Pseudomonas aeruginosa at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1AZU
 
 | |
1AUP
 
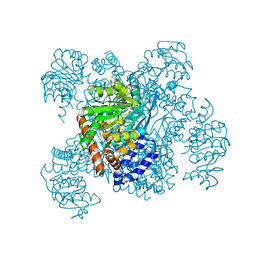 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | NAD-SPECIFIC GLUTAMATE DEHYDROGENASE | | Authors: | Baker, P.J, Waugh, M.L, Stillman, T.J, Turnbull, A.P, Rice, D.W. | | Deposit date: | 1997-09-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determinants of substrate specificity in the superfamily of amino acid dehydrogenases.
Biochemistry, 36, 1997
|
|
1BGV
 
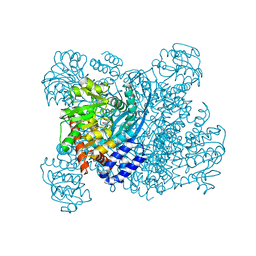 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE, GLUTAMIC ACID | | Authors: | Stillman, T.J, Baker, P.J, Britton, K.L, Rice, D.W. | | Deposit date: | 1998-06-01 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational flexibility in glutamate dehydrogenase. Role of water in substrate recognition and catalysis.
J.Mol.Biol., 234, 1993
|
|
3QH4
 
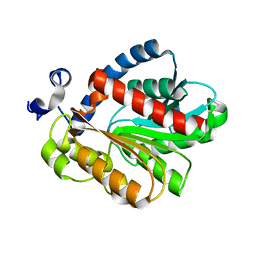 | |
3QHD
 
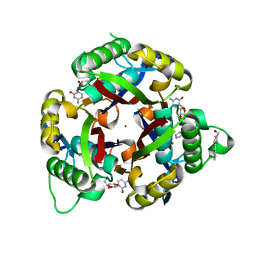 | |
3Q8H
 
 | | Crystal structure of 2c-methyl-d-erythritol 2,4-cyclodiphosphate synthase from burkholderia pseudomallei in complex with cytidine derivative EBSI01028 | | Descriptor: | 1,2-ETHANEDIOL, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 5'-deoxy-5'-[(imidazo[2,1-b][1,3]thiazol-5-ylcarbonyl)amino]cytidine, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2011-01-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cytidine derivatives as IspF inhibitors of Burkolderia pseudomallei.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3QHX
 
 | |
3RYY
 
 | | Fluoroalkyl and Alkyl Chains Have Similar Hydrophobicities in Binding to the Hydrophobic Wall of Carbonic Anhydrase | | Descriptor: | Carbonic anhydrase 2, N-propyl-4-sulfamoylbenzamide, ZINC ION | | Authors: | Snyder, P.W, Bai, S, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Fluoroalkyl and alkyl chains have similar hydrophobicities in binding to the "hydrophobic wall" of carbonic anhydrase.
J.Am.Chem.Soc., 133, 2011
|
|
3RZ8
 
 | | Fluoroalkyl and Alkyl Chains Have Similar Hydrophobicities in Binding to the Hydrophobic Wall of Carbonic Anhydrase | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, N-hexyl-4-sulfamoylbenzamide, ... | | Authors: | Snyder, P.W, Bai, S, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fluoroalkyl and alkyl chains have similar hydrophobicities in binding to the "hydrophobic wall" of carbonic anhydrase.
J.Am.Chem.Soc., 133, 2011
|
|
3RYZ
 
 | | Fluoroalkyl and Alkyl Chains Have Similar Hydrophobicities in Binding to the Hydrophobic Wall of Carbonic Anhydrase | | Descriptor: | Carbonic anhydrase 2, N-(2,2,3,3,4,4,4-heptafluorobutyl)-4-sulfamoylbenzamide, ZINC ION | | Authors: | Snyder, P.W, Bai, S, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Fluoroalkyl and alkyl chains have similar hydrophobicities in binding to the "hydrophobic wall" of carbonic anhydrase.
J.Am.Chem.Soc., 133, 2011
|
|
3RZ7
 
 | | Fluoroalkyl and Alkyl Chains Have Similar Hydrophobicities in Binding to the Hydrophobic Wall of Carbonic Anhydrase | | Descriptor: | 4-sulfamoyl-N-(2,2,3,3,4,4,5,5,6,6,6-undecafluorohexyl)benzamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Snyder, P.W, Bai, S, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fluoroalkyl and alkyl chains have similar hydrophobicities in binding to the "hydrophobic wall" of carbonic anhydrase.
J.Am.Chem.Soc., 133, 2011
|
|
3S72
 
 | | The origin of the hydrophobic effect in the molecular recognition of arylsulfonamides by carbonic anhydrase | | Descriptor: | 1H-benzimidazole-2-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Snyder, P.W, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanism of the hydrophobic effect in the biomolecular recognition of arylsulfonamides by carbonic anhydrase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3S78
 
 | |
3RYX
 
 | | Fluoroalkyl and Alkyl Chains Have Similar Hydrophobicities in Binding to the Hydrophobic Wall of Carbonic Anhydrase | | Descriptor: | Carbonic anhydrase 2, N-(2,2,3,3,3-pentafluoropropyl)-4-sulfamoylbenzamide, ZINC ION | | Authors: | Snyder, P.W, Bai, S, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fluoroalkyl and alkyl chains have similar hydrophobicities in binding to the "hydrophobic wall" of carbonic anhydrase.
J.Am.Chem.Soc., 133, 2011
|
|
3S77
 
 | | The origin of the hydrophobic effect in the molecular recognition of arylsulfonamides by carbonic anhydrase | | Descriptor: | 1,3-thiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Snyder, P.W, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Mechanism of the hydrophobic effect in the biomolecular recognition of arylsulfonamides by carbonic anhydrase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3RZ1
 
 | | Fluoroalkyl and Alkyl Chains Have Similar Hydrophobicities in Binding to the Hydrophobic Wall of Carbonic Anhydrase | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, N-(2,2,3,3,4,4,5,5,5-nonafluoropentyl)-4-sulfamoylbenzamide, ... | | Authors: | Snyder, P.W, Bai, S, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Fluoroalkyl and alkyl chains have similar hydrophobicities in binding to the "hydrophobic wall" of carbonic anhydrase.
J.Am.Chem.Soc., 133, 2011
|
|
3RZ5
 
 | | Fluoroalkyl and Alkyl Chains Have Similar Hydrophobicities in Binding to the Hydrophobic Wall of Carbonic Anhydrase | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, N-pentyl-4-sulfamoylbenzamide, ... | | Authors: | Snyder, P.W, Bai, S, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fluoroalkyl and alkyl chains have similar hydrophobicities in binding to the "hydrophobic wall" of carbonic anhydrase.
J.Am.Chem.Soc., 133, 2011
|
|
3T7C
 
 | |
3S74
 
 | |
3S75
 
 | |
3SX2
 
 | |
