6GME
 
 | |
1OWR
 
 | |
1P5Y
 
 | |
1OY2
 
 | | Coupling of Folding and Binding in the PTB Domain of the Signaling Protein Shc | | Descriptor: | SHC transforming protein | | Authors: | Farooq, A, Zeng, L, Yan, K.S, Ravichandran, K.S, Zhou, M.-M. | | Deposit date: | 2003-04-03 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Coupling of Folding and Binding in the PTB Domain of the Signaling Protein Shc
Structure, 11, 2003
|
|
5M7L
 
 | | Blastochloris viridis photosynthetic reaction center synchrotron structure | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL A, ... | | Authors: | Sharma, A.S, Johansson, L, Dunevall, E, Wahlgren, W.Y, Neutze, R, Katona, G. | | Deposit date: | 2016-10-28 | | Release date: | 2017-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Asymmetry in serial femtosecond crystallography data.
Acta Crystallogr A Found Adv, 73, 2017
|
|
6GNH
 
 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and an Azepanyl Phenyl Benzylsulphonamide Ligand | | Descriptor: | Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, methyl 4-(azepan-1-yl)-3-[(4-methoxyphenyl)sulfonylamino]benzoate | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
1OZI
 
 | | The alternatively spliced PDZ2 domain of PTP-BL | | Descriptor: | protein tyrosine phosphatase | | Authors: | Walma, T, Aelen, J, Oostendorp, M, van den Berk, L, Hendriks, W, Vuister, G.W. | | Deposit date: | 2003-04-09 | | Release date: | 2004-01-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Closed Binding Pocket and Global Destabilization Modify the Binding Properties of an Alternatively Spliced Form of the Second PDZ Domain of PTP-BL.
Structure, 12, 2004
|
|
6GLF
 
 | |
6GLL
 
 | | Crystal structure of hMTH1 N33A in complex with LW14 in the presence of acetate | | Descriptor: | 1~{H}-imidazo[4,5-b]pyridin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Eberle, S.A, Wiedmer, L, Sledz, P, Caflisch, A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | hMTH1 N33A in complex with LW14
To Be Published
|
|
4D76
 
 | | Human FXIa in complex with small molecule inhibitors. | | Descriptor: | COAGULATION FACTOR XI, GLYCEROL, N-[(2S)-1-({5-[(diaminomethylidene)amino]pentyl}amino)-1-oxo-3-phenylpropan-2-yl]-4-hydroxy-2-oxo-1,2-dihydroquinoline-6-carboxamide, ... | | Authors: | Sandmark, J, Oster, L, Jacso, T, Ullah, V, Redzick, A, Borjesson, U, Olsson, T, Norberg, M, Akerud, T. | | Deposit date: | 2014-11-20 | | Release date: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | From Mm Fragments to Nm Compounds Using Iloe-NMR to Guide Linking of Compounds in Fragment Based Drug Discovery (Fbdd).
To be Published
|
|
1P2Y
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME P450CAM IN COMPLEX WITH (S)-(-)-NICOTINE | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, Cytochrome P450-cam, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Strickler, M, Goldstein, B.M, Maxfield, K, Shireman, L, Kim, G, Matteson, D, Jones, J.P. | | Deposit date: | 2003-04-16 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic Studies on the Complex Behavior of Nicotine Binding to P450cam (CYP101)(dagger).
Biochemistry, 42, 2003
|
|
3TDL
 
 | | Structure of human serum albumin in complex with DAUDA | | Descriptor: | 11-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)undecanoic acid, MYRISTIC ACID, Serum albumin | | Authors: | Wang, Y, Luo, Z, Shi, X, Wang, H, Nie, L. | | Deposit date: | 2011-08-11 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A fluorescent fatty acid probe, DAUDA, selectively displaces two myristates bound in human serum albumin
Protein Sci., 20, 2011
|
|
5MBR
 
 | |
6GNV
 
 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and a isopropyl methyl indole aryl sulphonamide ligand | | Descriptor: | GLYCEROL, Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, ... | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
4CVN
 
 | | Structure of the Fap7-Rps14 complex | | Descriptor: | 30S RIBOSOMAL PROTEIN S11, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Loch, J, Blaud, M, Rety, S, Lebaron, S, Deschamps, P, Bareille, J, Jombart, J, Robert-Paganin, J, Delbos, L, Chardon, F, Zhang, E, Charenton, C, Tollervey, D, Leulliot, N. | | Deposit date: | 2014-03-28 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | RNA Mimicry by the Fap7 Adenylate Kinase in Ribosome Biogenesis
Plos Biol., 12, 2014
|
|
1P5W
 
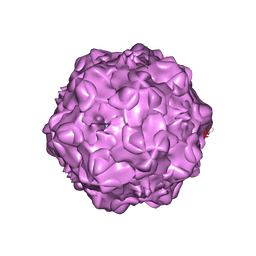 | |
8CEE
 
 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
1OJG
 
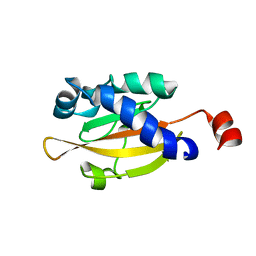 | | Sensory domain of the membraneous two-component fumarate sensor DcuS of E. coli | | Descriptor: | SENSOR PROTEIN DCUS | | Authors: | Pappalardo, L, Janausch, I.G, Vijayan, V, Zientz, E, Junker, J, Peti, W, Zweckstetter, M, Unden, G, Griesinger, C. | | Deposit date: | 2003-07-10 | | Release date: | 2003-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the sensory domain of the membranous two-component fumarate sensor (histidine protein kinase) DcuS of Escherichia coli.
J. Biol. Chem., 278, 2003
|
|
8CEC
 
 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CDU
 
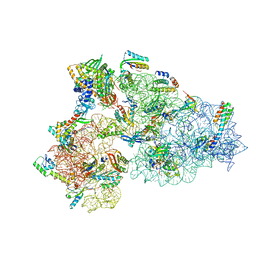 | | Rnase R bound to a 30S degradation intermediate (main state) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
1F7X
 
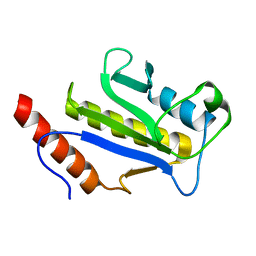 | | SOLUTION STRUCTURE OF C-TERMINAL DOMAIN ZIPA | | Descriptor: | CELL DIVISION PROTEIN ZIPA | | Authors: | Moy, F.J, Glasfeld, E, Mosyak, L, Powers, R. | | Deposit date: | 2000-06-28 | | Release date: | 2001-06-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ZipA, a crucial component of Escherichia coli cell division.
Biochemistry, 39, 2000
|
|
3TFW
 
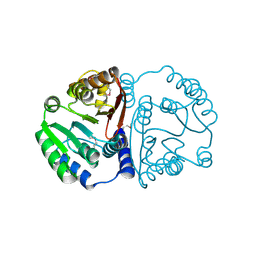 | |
8CED
 
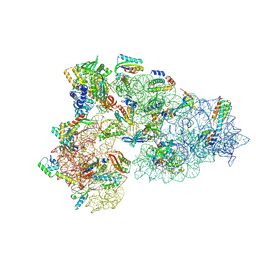 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
6GG1
 
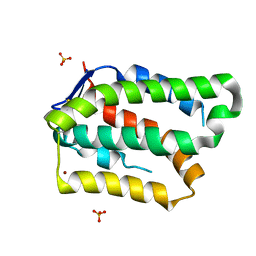 | | Structure of PROSS-edited human interleukin 24 | | Descriptor: | Interleukin-24, NICKEL (II) ION, SULFATE ION | | Authors: | Kolenko, P, Zahradnik, J, Kolarova, L, Schneider, B. | | Deposit date: | 2018-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Flexible regions govern promiscuous binding of IL-24 to receptors IL-20R1 and IL-22R1.
Febs J., 286, 2019
|
|
1OOY
 
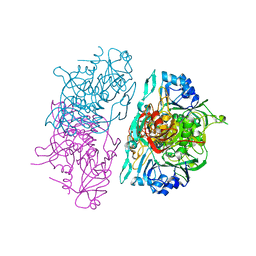 | | SUCCINYL-COA:3-KETOACID COA TRANSFERASE FROM PIG HEART | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Succinyl-CoA:3-ketoacid-coenzyme A transferase, ... | | Authors: | Coros, A.M, Swenson, L, Wolodko, W.T, Fraser, M.E. | | Deposit date: | 2003-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the CoA transferase from pig heart to 1.7 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
