3FNS
 
 | |
3FNT
 
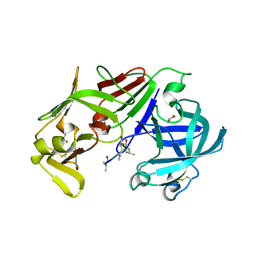 | | Crystal structure of pepstatin A bound histo-aspartic protease (HAP) from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, HAP protein, Inhibitor, ... | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2008-12-26 | | Release date: | 2009-05-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structures of the histo-aspartic protease (HAP) from plasmodium falciparum.
J.Mol.Biol., 388, 2009
|
|
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
4XTQ
 
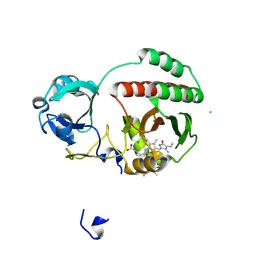 | | Crystal structure of a mutant (C20S) of a near-infrared fluorescent protein BphP1-FP | | Descriptor: | 3-[2-[(Z)-[5-[(Z)-[(3R,4R)-3-ethenyl-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, BphP1-FP/C20S, CHLORIDE ION | | Authors: | Pletnev, S, Malashkevich, V.N. | | Deposit date: | 2015-01-23 | | Release date: | 2015-12-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Molecular Basis of Spectral Diversity in Near-Infrared Phytochrome-Based Fluorescent Proteins.
Chem.Biol., 22, 2015
|
|
3QVC
 
 | |
3QVI
 
 | | Crystal structure of KNI-10395 bound histo-aspartic protease (HAP) from Plasmodium falciparum | | Descriptor: | (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-{[S-methyl-N-(phenylacetyl)-L-cysteinyl]ami no}-4-phenylbutanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-02-25 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the activation and inhibition of histo-aspartic protease from Plasmodium falciparum.
Biochemistry, 50, 2011
|
|
3QS1
 
 | | Crystal structure of KNI-10006 complex of Plasmepsin I (PMI) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Plasmepsin-1 | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-02-19 | | Release date: | 2011-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the free and inhibited forms of plasmepsin I (PMI) from Plasmodium falciparum.
J.Struct.Biol., 175, 2011
|
|
5XL6
 
 | |
5XL7
 
 | |
3QRV
 
 | |
5XLA
 
 | |
5XL4
 
 | |
5XLB
 
 | |
5XL8
 
 | |
5XL2
 
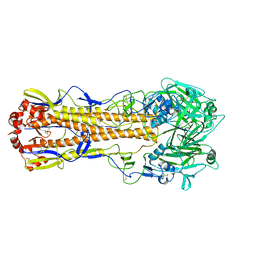 | |
9IN6
 
 | | Capsid of Vibrio cholerae phage mature VP1 | | Descriptor: | SHP of VP1, major capsid of VP1 | | Authors: | Liu, H.R, Pang, H. | | Deposit date: | 2024-07-05 | | Release date: | 2024-08-14 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Three-dimensional structures of Vibrio cholerae typing podophage VP1 in two states.
Structure, 32, 2024
|
|
9IOZ
 
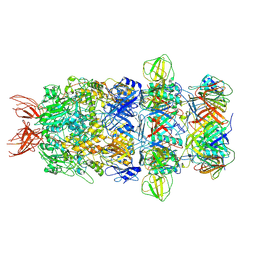 | | Structure of the bacteriophage T5 tail tip complex | | Descriptor: | Baseplate hub protein pb3, Baseplate tube protein p140, Distal tail protein pb9 | | Authors: | Peng, Y.N, Liu, H.R. | | Deposit date: | 2024-07-10 | | Release date: | 2024-09-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of Mature and Urea-Treated Empty Bacteriophage T5: Insights into Siphophage Infection and DNA Ejection.
Int J Mol Sci, 25, 2024
|
|
9ILV
 
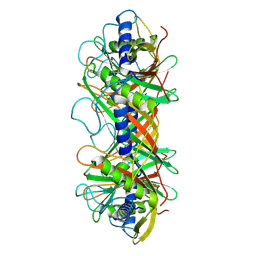 | | Structure of the bacteriophage T5 connector complex | | Descriptor: | Tail tube terminator protein p142 | | Authors: | Peng, Y.N, Liu, H.R. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structures of Mature and Urea-Treated Empty Bacteriophage T5: Insights into Siphophage Infection and DNA Ejection.
Int J Mol Sci, 25, 2024
|
|
9INY
 
 | | Structure of bacteriophage T5 tail tube | | Descriptor: | Tail tube protein pb6 | | Authors: | Peng, Y.N, Liu, H.R. | | Deposit date: | 2024-07-08 | | Release date: | 2024-09-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of Mature and Urea-Treated Empty Bacteriophage T5: Insights into Siphophage Infection and DNA Ejection.
Int J Mol Sci, 25, 2024
|
|
9ILP
 
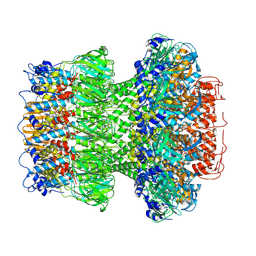 | | Structure of the bacteriophage T5 portal complex | | Descriptor: | Head completion protein, Portal protein pb7 | | Authors: | Peng, Y.N, Liu, H.R. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-18 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Mature and Urea-Treated Empty Bacteriophage T5: Insights into Siphophage Infection and DNA Ejection.
Int J Mol Sci, 25, 2024
|
|
9IMV
 
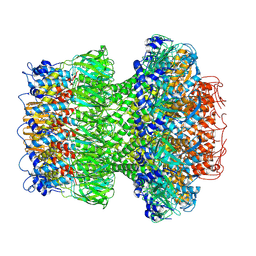 | |
9IMH
 
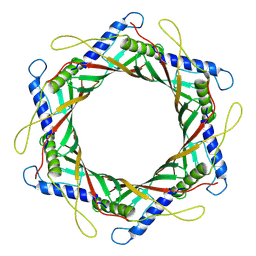 | |
9JZ0
 
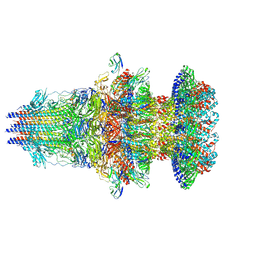 | | portal-tail complex of DNA-ejected T7 | | Descriptor: | Internal virion protein gp14, Portal protein, Protein 6.7, ... | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2024-10-13 | | Release date: | 2024-12-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational changes in and translocation of small proteins: insights into the ejection mechanism of podophages.
J.Virol., 99, 2025
|
|
9JYY
 
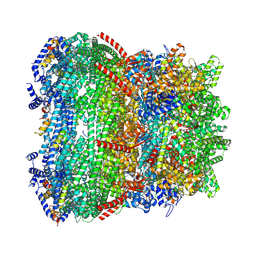 | | core proteins of mature T7 | | Descriptor: | Internal virion protein gp14, Internal virion protein gp15, Peptidoglycan transglycosylase gp16, ... | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2024-10-13 | | Release date: | 2024-12-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational changes in and translocation of small proteins: insights into the ejection mechanism of podophages.
J.Virol., 99, 2025
|
|
9JYZ
 
 | | portal-tail complex of mature T7 | | Descriptor: | Portal protein, Protein 6.7, Protein 7.3, ... | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2024-10-13 | | Release date: | 2024-12-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Conformational changes in and translocation of small proteins: insights into the ejection mechanism of podophages.
J.Virol., 99, 2025
|
|
