8AMS
 
 | | Complex of human TRIM2 RING domain, UBCH5C, and Ubiquitin | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, GLYCEROL, Polyubiquitin-C, ... | | Authors: | Perez-Borrajero, C, Kotova, I, Murciano, B, Hennig, J. | | Deposit date: | 2022-08-04 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biophysical studies of TRIM2 and TRIM3
To Be Published
|
|
8B24
 
 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 3600-seconds post reaction initiation with Na+ | | Descriptor: | DIPHOSPHATE, K(+)-stimulated pyrophosphate-energized sodium pump, MAGNESIUM ION, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (4.53 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
8B22
 
 | |
8B23
 
 | |
8B21
 
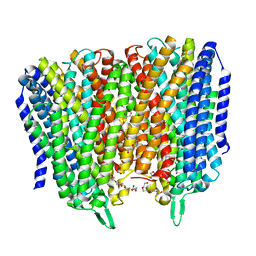 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 0-60-seconds post reaction initiation with Na+ | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-BETA-D-MALTOSIDE, K(+)-stimulated pyrophosphate-energized sodium pump, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
7R7M
 
 | |
7R3U
 
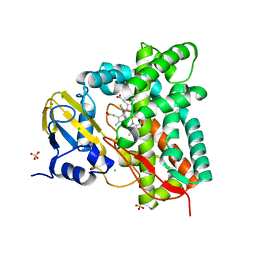 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | 1-[4-(1,2,3-thiadiazol-4-yl)phenyl]methanamine, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Snee, M, Katariya, M, Leys, D, Levy, C. | | Deposit date: | 2022-02-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
7RVA
 
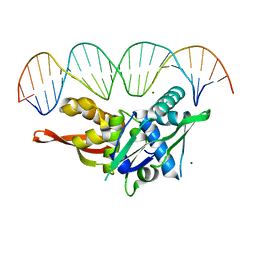 | | Updated Crystal Structure of Replication Initiator Protein REPE54. | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*T)-3'), DNA (5'-D(*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*T)-3'), MAGNESIUM ION, ... | | Authors: | Ward, A.R, Snow, C.D. | | Deposit date: | 2021-08-18 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Stabilizing DNA-Protein Co-Crystals via Intra-Crystal Chemical Ligation of the DNA
Crystals, 12, 2022
|
|
7SDP
 
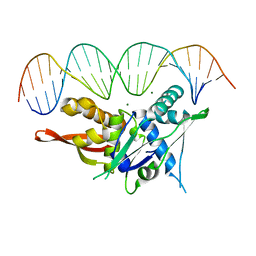 | |
7SGC
 
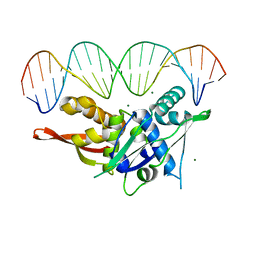 | |
7SPM
 
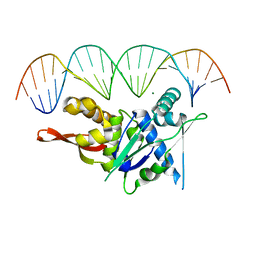 | | Replication Initiator Protein REPE54 and cognate DNA sequence with terminal three prime phosphates chemically crosslinked (30 mg/mL EDC, 12 hours, 2 doses). | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*A)-3'), DNA (5'-D(*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Ward, A.R, Snow, C.D. | | Deposit date: | 2021-11-02 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Stabilizing DNA-Protein Co-Crystals via Intra-Crystal Chemical Ligation of the DNA
Crystals, 12, 2022
|
|
7SOZ
 
 | | Replication Initiator Protein REPE54 and cognate DNA sequence with terminal three prime phosphates chemically crosslinked (5 mg/mL EDC, 12 hours). | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*A)-3'), DNA (5'-D(*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Ward, A.R, Snow, C.D. | | Deposit date: | 2021-11-01 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
7SQM
 
 | | Discovery and Preclinical Pharmacology of INE963, A Potent and Fast-Acting Blood-Stage Antimalarial with a High Barrier to Resistance and Potential for Single-Dose Cure in Uncomplicated Malaria | | Descriptor: | 1-[(4S)-5-(2,4-difluorophenyl)imidazo[2,1-b][1,3,4]thiadiazol-2-yl]-4-methylpiperidin-4-amine, GLYCEROL, Serine/threonine-protein kinase haspin | | Authors: | Shu, W, Yokokawa, F. | | Deposit date: | 2021-11-05 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Failure of artesunate-mefloquine combination therapy for uncompli-cated Plasmodium falciparum malaria in southern Cambodia.
Malar. J., 2009, 8, 10, 2009
|
|
7T7U
 
 | | Light Harvesting complex phycocyanin PC 630, from the cryptophyte Chroomonas sp. M1627 | | Descriptor: | DiCys-(15,16)-Dihydrobiliverdin, GLYCEROL, PHYCOCYANOBILIN, ... | | Authors: | Michie, K.A, Harrop, S.J, Rathbone, H.W, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2021-12-15 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular structures reveal the origin of spectral variation in cryptophyte light harvesting antenna proteins.
Protein Sci., 32, 2023
|
|
7T89
 
 | | Light harvesting complex Phycocyanin PC577 from the cryptophyte Hemiselmis pacifica CCMP 706 | | Descriptor: | DiCys-(15,16)-Dihydrobiliverdin, PHYCOCYANOBILIN, Phycoerythrin alpha subunit 1, ... | | Authors: | Michie, K.A, Curmi, P.C, Harrop, S, Rathbone, H.W. | | Deposit date: | 2021-12-16 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Molecular structures reveal the origin of spectral variation in cryptophyte light harvesting antenna proteins.
Protein Sci., 32, 2023
|
|
7S1G
 
 | | wild-type Escherichia coli stalled ribosome with antibiotic linezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S1H
 
 | | Wild-type Escherichia coli ribosome with antibiotic linezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S86
 
 | |
7S7S
 
 | |
8OVR
 
 | |
8OWF
 
 | | Clostridium perfringens chitinase CP4_3455 with chitosan | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Chitodextrinase, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-04-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Clostridium perfringens chitinase CP4_3455 with chitosan
To Be Published
|
|
8OXU
 
 | | Crystal Structure of the Hsp90-LA1011 Complex | | Descriptor: | ATP-dependent molecular chaperone HSP82, dimethyl 2,6-bis[2-(dimethylamino)ethyl]-1-methyl-4-[4-(trifluoromethyl)phenyl]-4~{H}-pyridine-3,5-dicarboxylate | | Authors: | Roe, S.M, Prodromou, C. | | Deposit date: | 2023-05-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | The Crystal Structure of the Hsp90-LA1011 Complex and the Mechanism by Which LA1011 May Improve the Prognosis of Alzheimer's Disease.
Biomolecules, 13, 2023
|
|
8PFF
 
 | | Galectin-3C in complex with a triazolesulfone derivative | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-(phenylsulfonyl)-1,2,3-triazol-1-yl]oxan-2-yl]sulfanyl-oxane-3,4,5-triol, Galectin-3, MAGNESIUM ION, ... | | Authors: | Kumar, R, Mahanti, M, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Ligand Sulfur Oxidation State Progressively Alters Galectin-3-Ligand Complex Conformations To Induce Affinity-Influencing Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
8OV8
 
 | | Crystal structure of Ene-reductase 1 from black poplar mushroom in complex to NADP | | Descriptor: | Ene-reductase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, ... | | Authors: | Korf, L, Essen, L.-O, Karrer, D, Ruehl, M. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Shifting the substrate scope of an ene/yne-reductase by loop engineering
To Be Published
|
|
8QI7
 
 | |
