8OQK
 
 | | Crystal structure of Tannerella forsythia sugar kinase K1058 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-acetylglucosamine kinase | | Authors: | Gogler, K, Fink, P, Stasiak, A.C, Stehle, T, Zocher, G. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
5CPU
 
 | | Crystal structure of murine polyomavirus PTA strain VP1 | | Descriptor: | GLYCEROL, VP1 | | Authors: | Liaci, A.M, Buch, M.H.C, Neu, U, Stehle, T. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity.
Plos Pathog., 11, 2015
|
|
5CQ0
 
 | | Crystal structure of murine polyomavirus RA strain VP1 in complex with the GD1a glycan | | Descriptor: | Capsid protein VP1, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose, ... | | Authors: | Buch, M.H.C, Liaci, A.M, Neu, U, Stehle, T. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity.
Plos Pathog., 11, 2015
|
|
1U8C
 
 | | A novel adaptation of the integrin PSI domain revealed from its crystal structure | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, J.P, Stehle, T, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2004-08-05 | | Release date: | 2004-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A novel adaptation of the integrin PSI domain revealed from its crystal structure.
J.Biol.Chem., 279, 2004
|
|
5CPZ
 
 | | Crystal structure of murine polyomavirus RA strain VP1 in complex with the GT1a glycan | | Descriptor: | Capsid protein VP1, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose, ... | | Authors: | Buch, M.H.C, Liaci, A.M, Neu, U, Stehle, T. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity.
Plos Pathog., 11, 2015
|
|
5CPY
 
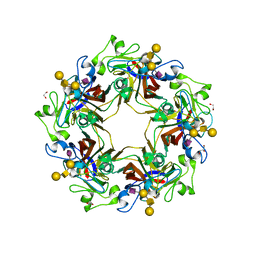 | | Crystal structure of murine polyomavirus PTA strain VP1 in complex with the GD1a glycan | | Descriptor: | GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose, VP1 | | Authors: | Liaci, A.M, Buch, M.H.C, Neu, U, Stehle, T. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity.
Plos Pathog., 11, 2015
|
|
5CPW
 
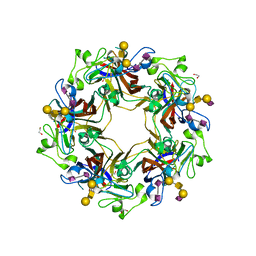 | | Crystal structure of murine polyomavirus PTA strain VP1 in complex with the GT1a glycan | | Descriptor: | GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose, ... | | Authors: | Liaci, A.M, Buch, M.H.C, Neu, U, Stehle, T. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity.
Plos Pathog., 11, 2015
|
|
1JV2
 
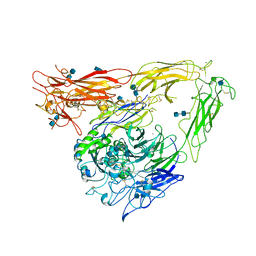 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR SEGMENT OF INTEGRIN ALPHAVBETA3 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, J.P, Stehle, T, Diefenbach, B, Zhang, R, Dunker, R, Scott, D, Joachimiak, A, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2001-08-28 | | Release date: | 2001-10-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the extracellular segment of integrin alpha Vbeta3.
Science, 294, 2001
|
|
1KKE
 
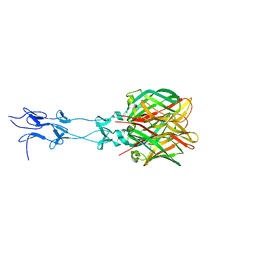 | |
7B6U
 
 | |
7B6S
 
 | | Sheep Polyomavirus VP1 in complex with 10 mM Forssman antigen pentaose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose, ... | | Authors: | Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-12-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | A novel and highly specific Forssman antigen-binding protein from sheep polyomavirus
Biorxiv, 2023
|
|
7B6T
 
 | | Sheep Polyomavirus VP1 in complex with 10 mM globo-N-tetraose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose, ... | | Authors: | Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-12-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | A novel and highly specific Forssman antigen-binding protein from sheep polyomavirus
Biorxiv, 2023
|
|
7B6V
 
 | | Sheep Polyomavirus VP1 in complex with 5 mM Forssman antigen pentaose and 20 mM 3'-sialyllactosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose, 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-12-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | A novel and highly specific Forssman antigen-binding protein from sheep polyomavirus
Biorxiv, 2023
|
|
1IC1
 
 | | THE CRYSTAL STRUCTURE FOR THE N-TERMINAL TWO DOMAINS OF ICAM-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, INTERCELLULAR ADHESION MOLECULE-1 | | Authors: | Casasnovas, J.M, Stehle, T, Liu, J.-H, Wang, J.-H, Springer, T.A. | | Deposit date: | 1998-03-09 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A dimeric crystal structure for the N-terminal two domains of intercellular adhesion molecule-1.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
3S3L
 
 | | Crystal Structure of CerJ from Streptomyces tendae | | Descriptor: | ACETATE ION, CHLORIDE ION, CerJ | | Authors: | Zocher, G, Hertweck, C, Bretschneider, T, Stehle, T. | | Deposit date: | 2011-05-18 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
1L5G
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR SEGMENT OF INTEGRIN AVB3 IN COMPLEX WITH AN ARG-GLY-ASP LIGAND | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, J.-P, Stehle, T, Zhang, R, Joachimiak, A, Frech, M, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2002-03-06 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand.
Science, 296, 2002
|
|
3T6S
 
 | | Crystal structure of CerJ from Streptomyces tendae in Complex with CoA | | Descriptor: | COENZYME A, CerJ | | Authors: | Zocher, G, Bretschneider, T, Hertweck, C, Stehle, T. | | Deposit date: | 2011-07-29 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
3T5Y
 
 | | Crystal structure of CerJ from Streptomyces tendae - malonic acid covalently linked to the catalytic Cystein C116 | | Descriptor: | ACETATE ION, CerJ | | Authors: | Zocher, G, Bretschneider, T, Hertweck, C, Stehle, T. | | Deposit date: | 2011-07-28 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
3T8E
 
 | | Crystal structure of CerJ from Streptomyces tendae soaked with CerviK | | Descriptor: | ACETATE ION, CerJ | | Authors: | Zocher, G, Bretschneider, T, Hertweck, C, Stehle, T. | | Deposit date: | 2011-08-01 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
1LY2
 
 | | Crystal structure of unliganded human CD21 SCR1-SCR2 (Complement receptor type 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, complement receptor type 2 | | Authors: | Prota, A.E, Sage, D.R, Stehle, T, Fingeroth, J.D. | | Deposit date: | 2002-06-06 | | Release date: | 2002-07-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of human CD21: Implications for Epstein-Barr virus and C3d binding.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1M1X
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR SEGMENT OF INTEGRIN ALPHA VBETA3 BOUND TO MN2+ | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, J.-P, Stehle, T, Zhang, R, Joachimiak, A, Frech, M, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand.
Science, 296, 2002
|
|
4MJ0
 
 | | BK Polyomavirus VP1 pentamer in complex with GD3 oligosaccharide | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid, ... | | Authors: | Neu, U, Stroh, L.J, Stehle, T. | | Deposit date: | 2013-09-03 | | Release date: | 2013-11-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Structure-Guided Mutation in the Major Capsid Protein Retargets BK Polyomavirus.
Plos Pathog., 9, 2013
|
|
3S6Y
 
 | | Structure of reovirus attachment protein sigma1 in complex with alpha-2,6-sialyllactose | | Descriptor: | N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Outer capsid protein sigma-1 | | Authors: | Reiter, D.M, Dermody, T.S, Stehle, T. | | Deposit date: | 2011-05-26 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of reovirus attachment protein sigma1 in complex with sialylated oligosaccharides
Plos Pathog., 7, 2011
|
|
4E0T
 
 | | Crystal structure of CdpNPT in its unbound state | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cyclic dipeptide N-prenyltransferase, ... | | Authors: | Schuller, J.M, Zocher, G, Stehle, T. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and catalytic mechanism of a cyclic dipeptide prenyltransferase with broad substrate promiscuity.
J.Mol.Biol., 422, 2012
|
|
4EE6
 
 | |
