9AX2
 
 | | Structure of full-length amyloidogenic immunoglobulin light chain H9 in complex with 2-(1-methyl-3-oxo-5-(2-phenylpropoxy)isoindolin-2-yl)ethyl (3-(1H-imidazol-4-yl)benzyl)carbamate | | Descriptor: | 2-{(1R)-1-methyl-3-oxo-5-[(2S)-2-phenylpropoxy]-1,3-dihydro-2H-isoindol-2-yl}ethyl {[(3M)-3-(1H-imidazol-4-yl)phenyl]methyl}carbamate, H9 Immunoglobulin Light Chain, PHOSPHATE ION | | Authors: | Lederberg, O.L, Yan, N.L, Stanfield, R.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2024-03-05 | | Release date: | 2024-12-18 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of Potent and Selective Pyridone-Based Small Molecule Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains.
J.Med.Chem., 67, 2024
|
|
9AX3
 
 | | Structure of full-length amyloidogenic immunoglobulin light chain H9 in complex with 4-(3-ethyl-3-phenylpyrrolidin-1-yl)-1,6-dimethylpyridin-2(1H)-one | | Descriptor: | 4-[(3R)-3-ethyl-3-phenylpyrrolidin-1-yl]-1,6-dimethylpyridin-2(1H)-one, H9 Immunoglobulin Light Chain, PHOSPHATE ION | | Authors: | Lederberg, O.L, Yan, N.L, Stanfield, R.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2024-03-05 | | Release date: | 2024-12-18 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Potent and Selective Pyridone-Based Small Molecule Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains.
J.Med.Chem., 67, 2024
|
|
9AWY
 
 | | Structure of full-length amyloidogenic immunoglobulin light chain H9 in complex with 3-(2-(4-(3-ethyl-3-phenylpyrrolidin-1-yl)-6-methyl-2-oxopyridin-1(2H)-yl)ethyl)-8-(imidazo[1,5-a]pyrazin-8-yl)-1,3,8-triazaspiro[4.5]decan-2-one | | Descriptor: | 3-(2-{4-[(3R)-3-ethyl-3-phenylpyrrolidin-1-yl]-6-methyl-2-oxopyridin-1(2H)-yl}ethyl)-8-[(4R)-imidazo[1,5-a]pyrazin-8-yl]-1,3,8-triazaspiro[4.5]decan-2-one, H9 Immunoglobulin Light Chain, PHOSPHATE ION | | Authors: | Lederberg, O.L, Kelly, J.W, Yan, N.L, Wilson, I.A, Stanfield, R.L. | | Deposit date: | 2024-03-05 | | Release date: | 2024-12-18 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Potent and Selective Pyridone-Based Small Molecule Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains.
J.Med.Chem., 67, 2024
|
|
9AWX
 
 | | Structure of full-length amyloidogenic immunoglobulin light chain H9 in complex with 1-(1-(imidazo[1,5-a]pyrazin-8-yl)azetidin-3-yl)-3-(2-(6-methyl-4-(3-methyl-3-phenylpyrrolidin-1-yl)-2-oxopyridin-1(2H)-yl)ethyl)urea | | Descriptor: | H9 Immunoglobulin Light Chain, N-{1-[(4R)-imidazo[1,5-a]pyrazin-8-yl]azetidin-3-yl}-N'-(2-{6-methyl-4-[(3R)-3-methyl-3-phenylpyrrolidin-1-yl]-2-oxopyridin-1(2H)-yl}ethyl)urea, PHOSPHATE ION | | Authors: | Lederberg, O.L, Yan, N.L, Stanfield, R.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2024-03-05 | | Release date: | 2024-12-18 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of Potent and Selective Pyridone-Based Small Molecule Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains.
J.Med.Chem., 67, 2024
|
|
9AX1
 
 | | Structure of full-length amyloidogenic immunoglobulin light chain H9 in complex with 2-(6-methyl-2-oxo-4-(2-phenylpropoxy)pyridin-1(2H)-yl)ethyl (3-(1H-imidazol-4-yl)benzyl)carbamate | | Descriptor: | 2-{6-methyl-2-oxo-4-[(2S)-2-phenylpropoxy]pyridin-1(2H)-yl}ethyl {[(3P)-3-(1H-imidazol-4-yl)phenyl]methyl}carbamate, H9 Immunoglobulin Light Chain, PHOSPHATE ION | | Authors: | Lederberg, O.L, Yan, N.L, Stanfield, R.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2024-03-05 | | Release date: | 2024-12-18 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Potent and Selective Pyridone-Based Small Molecule Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains.
J.Med.Chem., 67, 2024
|
|
6J7O
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant E146Q from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
6J7T
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant D82A from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
6J7R
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant S78A co-expressed with TakA from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
6J7N
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant D82A co-expressed with TakA from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
6J7S
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) wild type protein from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
6J7P
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant E146Q co-expressed with TakA from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.629 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
6J7Q
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant S78A from Mycobacterium tuberculosis | | Descriptor: | CALCIUM ION, MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
7CDW
 
 | |
7BL6
 
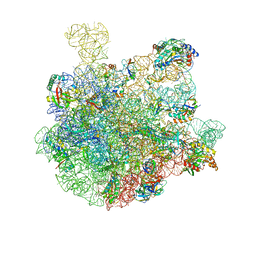 | | 50S-ObgE-GMPPNP particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Hilal, T, Nikolay, R, Schmidt, S, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7BL2
 
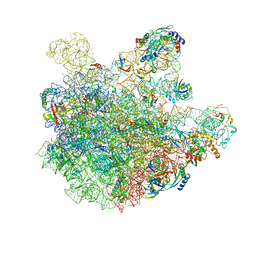 | | pre-50S-ObgE particle state 1 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Hilal, T, Nikolay, R, Schmidt, S, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7BL3
 
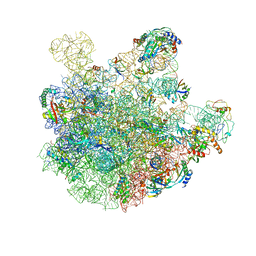 | | pre-50S-ObgE particle state 2 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Hilal, T, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7BL5
 
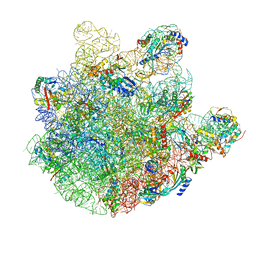 | | pre-50S-ObgE particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Hilal, T, Nikolay, R, Spahn, C.M.T, Schmidt, S. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7BL4
 
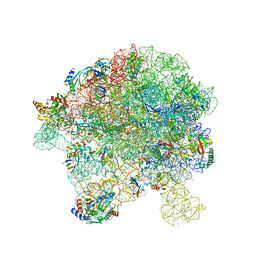 | | in vitro reconstituted 50S-ObgE-GMPPNP-RsfS particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Hilal, T, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7EO7
 
 | | Crystal structure of HCoV-NL63 3C-like protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24916625 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7EO8
 
 | | Crystal structure of SARS coronavirus main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2808516 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7DV4
 
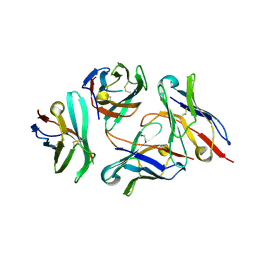 | | Crystal structure of anti-CTLA-4 VH domain in complex with human CTLA-4 | | Descriptor: | 1,2-ETHANEDIOL, 4003-1(VH), Cytotoxic T-lymphocyte protein 4 | | Authors: | Li, H, Gan, X, He, Y. | | Deposit date: | 2021-01-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | An anti-CTLA-4 heavy chain-only antibody with enhanced T reg depletion shows excellent preclinical efficacy and safety profile.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7CMD
 
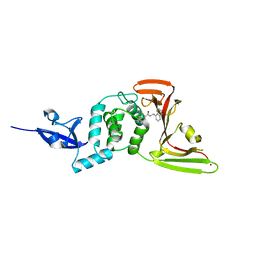 | | Crystal structure of the SARS-CoV-2 PLpro with GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, ZINC ION | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of SARS-CoV-2 papain-like protease.
Acta Pharm Sin B, 11, 2021
|
|
7CJD
 
 | |
