8IWA
 
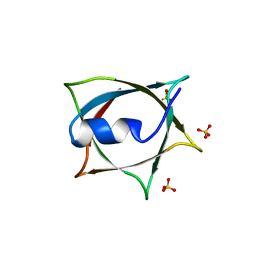 | | Crystal structure of Q9PR55 at pH 6.5 | | Descriptor: | SULFATE ION, Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8IWB
 
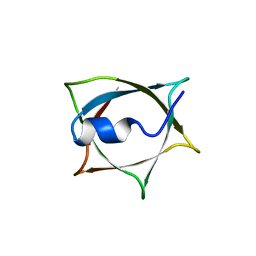 | | Crystal structure of Q9PR55 at pH 7.5 | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8HW9
 
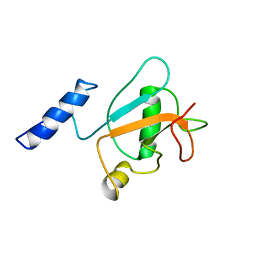 | | Solution structure of ubiquitin-like domain (UBL) of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1 | | Authors: | Lai, C.H, Ko, K.T, Fan, P.J, Yu, T.A, Chang, C.F, Hsu, S.T.D. | | Deposit date: | 2022-12-29 | | Release date: | 2024-01-31 | | Last modified: | 2024-08-14 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the ZFAND1-p97 interaction involved in stress granule clearance.
J.Biol.Chem., 300, 2024
|
|
7VPW
 
 | |
7F63
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-45 (Focused refinement of S-RBD and chAb-45 region) | | Descriptor: | RBD-chAb45, Heavy chain, Light chain, ... | | Authors: | Yang, T.J, Yu, P.Y, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-06-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure-guided antibody cocktail for prevention and treatment of COVID-19.
Plos Pathog., 17, 2021
|
|
7F62
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-25 (Focused refinement of S-RBD and chAb-25 region) | | Descriptor: | RBD-chAb-25, Heavy chain, Light chain, ... | | Authors: | Yang, T.J, Yu, P.Y, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-06-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-guided antibody cocktail for prevention and treatment of COVID-19.
Plos Pathog., 17, 2021
|
|
6AHW
 
 | |
5ZYO
 
 | |
7EB0
 
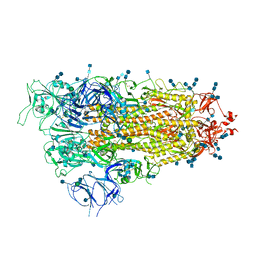 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, one RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
7EAZ
 
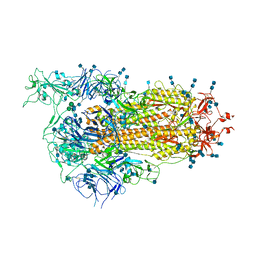 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, one RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
7EB5
 
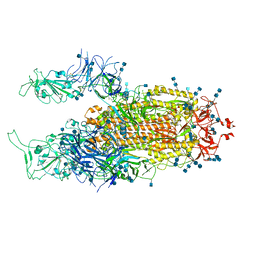 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, two RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
7EB3
 
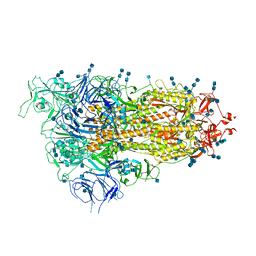 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, one RBD-up conformation 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
7EB4
 
 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, two RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
7EH5
 
 | | Cryo-EM structure of SARS-CoV-2 S-D614G variant in complex with neutralizing antibodies, RBD-chAb15 and RBD-chAb45 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RBD-chAb15, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-03-28 | | Release date: | 2021-09-01 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7EDJ
 
 | | Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2 (ACE2) ectodomain, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7EDI
 
 | | Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7), two RBD-up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-01 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7EDH
 
 | | Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7), one RBD-up conformation 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-01 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7EDF
 
 | | Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7), one RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-01 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7EDG
 
 | | Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7), one RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function.
Nat.Struct.Mol.Biol., 28, 2021
|
|
