6MD1
 
 | |
2FK5
 
 | | Crystal structure of l-fuculose-1-phosphate aldolase from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, SULFATE ION, fuculose-1-phosphate aldolase | | Authors: | Jeyakanthan, J, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-04 | | Release date: | 2007-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Purification, crystallization and preliminary X-ray crystallographic study of the L-fuculose-1-phosphate aldolase (FucA) from Thermus thermophilus HB8
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2FLF
 
 | | Crystal structure of l-fuculose-1-phosphate aldolase from Thermus Thermophilus HB8 | | Descriptor: | fuculose-1-phosphate aldolase | | Authors: | Jeyakanthan, J, Yokoyama, S, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-06 | | Release date: | 2007-01-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Purification, crystallization and preliminary X-ray crystallographic study of the L-fuculose-1-phosphate aldolase (FucA) from Thermus thermophilus HB8
Acta Crystallogr.,Sect.F, 61, 2005
|
|
8FHE
 
 | |
8FHF
 
 | |
8FHG
 
 | |
5ZY9
 
 | | Structural basis for a tRNA synthetase | | Descriptor: | (1R,2R)-2-[(2S,4E,6E,8R,9S,11R,13S,15S,16S)-7-cyano-8,16-dihydroxy-9,11,13,15-tetramethyl-18-oxooxacyclooctadeca-4,6-dien-2-yl]cyclopentanecarboxylic acid, Threonyl-tRNA synthase, ZINC ION | | Authors: | Zhang, J. | | Deposit date: | 2018-05-23 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for inhibition of
translational functions on a tRNA synthetase
To Be Published
|
|
8TDM
 
 | | Cryo-EM structure of AtMSL10-K539E | | Descriptor: | Mechanosensitive ion channel protein 10 | | Authors: | Zhang, J, Yuan, P. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Open structure and gating of the Arabidopsis mechanosensitive ion channel MSL10.
Nat Commun, 14, 2023
|
|
8TDJ
 
 | |
8TDK
 
 | | Cryo-EM structure of AtMSL10-G556V | | Descriptor: | Mechanosensitive ion channel protein 10 | | Authors: | Zhang, J, Yuan, P. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Open structure and gating of the Arabidopsis mechanosensitive ion channel MSL10.
Nat Commun, 14, 2023
|
|
8TDL
 
 | |
8GSJ
 
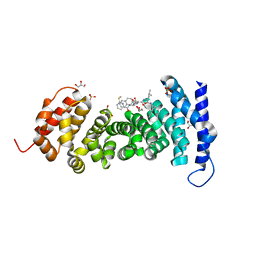 | | APC-Asef tripeptide inhibitor | | Descriptor: | (1R,2S)-2-phenylcyclopropanamine, 2-methylsulfanylpyrimidine-4-carbaldehyde, Adenomatous polyposis coli protein, ... | | Authors: | Zhang, J, Wang, X.F, Song, K. | | Deposit date: | 2022-09-06 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | APC-Asef tripeptide inhibitor
To Be Published
|
|
3JBU
 
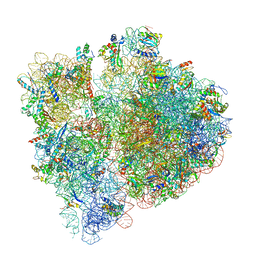 | | Mechanisms of Ribosome Stalling by SecM at Multiple Elongation Steps | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, J, Pan, X.J, Yan, K.G, Sun, S, Gao, N, Sui, S.F. | | Deposit date: | 2015-10-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Mechanisms of ribosome stalling by SecM at multiple elongation steps
Elife, 4, 2015
|
|
6AR6
 
 | |
6AUG
 
 | |
6LZ1
 
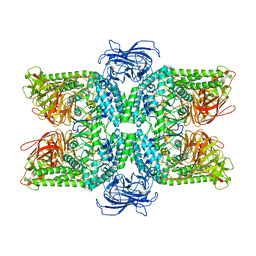 | | Structure of S.pombe alpha-mannosidase Ams1 | | Descriptor: | Ams1, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of fission yeast tetrameric alpha-mannosidase Ams1.
Febs Open Bio, 10, 2020
|
|
6AVI
 
 | |
3JBV
 
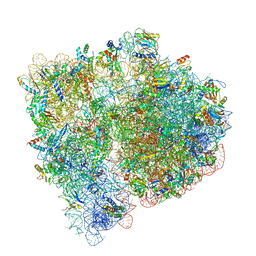 | | Mechanisms of Ribosome Stalling by SecM at Multiple Elongation Steps | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Zhang, J, Pan, X.J, Yan, K.G, Sun, S, Gao, N, Sui, S.F. | | Deposit date: | 2015-10-16 | | Release date: | 2016-01-27 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Mechanisms of ribosome stalling by SecM at multiple elongation steps
Elife, 4, 2015
|
|
4TZW
 
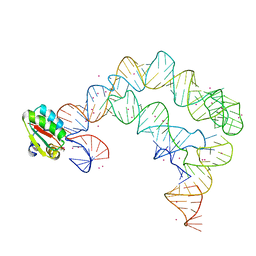 | | Co-crystals of the Ternary Complex Containing a T-box Stem I RNA, its Cognate tRNAGly, and B. subtilis YbxF protein, treated by removing lithium sulfate and replacing Mg2+ with Sr2+ post crystallization | | Descriptor: | Ribosome-associated protein L7Ae-like, STRONTIUM ION, T-box Stem I, ... | | Authors: | Zhang, J, Ferre-D'Amare, A.R. | | Deposit date: | 2014-07-11 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.671 Å) | | Cite: | Dramatic Improvement of Crystals of Large RNAs by Cation Replacement and Dehydration.
Structure, 22, 2014
|
|
4TZP
 
 | | As Grown, Untreated Co-crystals of the Ternary Complex Containing a T-box Stem I RNA, its cognate tRNAGly, and B. subtilis YbxF protein | | Descriptor: | Ribosome-associated protein L7Ae-like, engineered tRNA, glyQ T-box Stem I | | Authors: | Zhang, J, Ferre-D'Amare, A.R. | | Deposit date: | 2014-07-10 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (8.503 Å) | | Cite: | Dramatic Improvement of Crystals of Large RNAs by Cation Replacement and Dehydration.
Structure, 22, 2014
|
|
4TZV
 
 | | Co-crystals of the Ternary Complex Containing a T-box Stem I RNA, its Cognate tRNAGly, and B. subtilis YbxF protein, treated by removing lithium sulfate post crystallization | | Descriptor: | Ribosome-associated protein L7Ae-like, T-box stem I, engineered tRNA | | Authors: | Zhang, J, Ferre-D'Amare, A.R. | | Deposit date: | 2014-07-11 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (5.03 Å) | | Cite: | Dramatic Improvement of Crystals of Large RNAs by Cation Replacement and Dehydration.
Structure, 22, 2014
|
|
4G3H
 
 | | Crystal structure of helicobacter pylori arginase | | Descriptor: | Arginase (RocF), MANGANESE (II) ION | | Authors: | Zhang, J, Zhang, X, Li, D, Hu, Y, Zou, Q, Wang, D. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function studies on Helicobacter pylori arginase
To be Published
|
|
1SPF
 
 | |
7VQO
 
 | | Cryo-EM structure of Ams1 bound to the FW domain of Nbr1 | | Descriptor: | Ams1, Nbr1 and malE fusion protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2021-10-20 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural mechanism of protein recognition by the FW domain of autophagy receptor Nbr1
Nat Commun, 13, 2022
|
|
7KRR
 
 | | Structural impact on SARS-CoV-2 spike protein by D614G substitution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Lu, J.M, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Zhu, H.S, Woosley, A.N, Yang, W, Sliz, P, Chen, B. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-24 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural impact on SARS-CoV-2 spike protein by D614G substitution.
Science, 372, 2021
|
|
