3GR7
 
 | |
8A8I
 
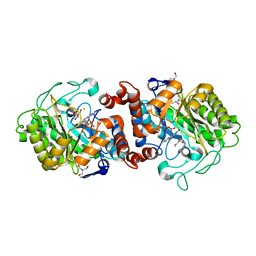 | | Xenobiotic reductase A from Pseudomonas putida in complex with ethyl (Z)-2-(hydroxyimino)-3-oxobutanoate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Polidori, N, Gruber, K. | | Deposit date: | 2022-06-23 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mechanistic Insights into the Ene-Reductase-Catalyzed Promiscuous Reduction of Oximes to Amines.
Acs Catalysis, 13, 2023
|
|
8ACS
 
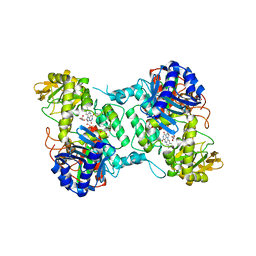 | | Crystal structure of FMO from Janthinobacterium svalbardensis | | Descriptor: | DI(HYDROXYETHYL)ETHER, FAD-dependent oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Polidori, N, Galuska, P, Gruber, K. | | Deposit date: | 2022-07-06 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Cold-Active Flavin-Dependent Monooxygenase from Janthinobacterium svalbardensis Unlocks Applications of Baeyer-Villiger Monooxygenases at Low Temperature.
Acs Catalysis, 13, 2023
|
|
8AUL
 
 | |
8AWP
 
 | |
8AU9
 
 | |
8AUF
 
 | |
8AU8
 
 | |
8AUG
 
 | |
8AUN
 
 | |
8AUM
 
 | |
8AUQ
 
 | |
8AUO
 
 | |
8AUI
 
 | |
8AUE
 
 | |
8AUJ
 
 | |
8AUH
 
 | |
8AUB
 
 | |
8AWN
 
 | | Crystal structure of a manganese-containing cupin (tm1459) from Thermotoga maritima, variant C106Q | | Descriptor: | CHLORIDE ION, Cupin_2 domain-containing protein | | Authors: | Grininger, C, Steiner, K, Gruber, K, Pavkov-Keller, T. | | Deposit date: | 2022-08-30 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Engineering TM1459 for Stabilisation against Inactivation by Amino Acid Oxidation
Chem Ing Tech, 2023
|
|
8AUA
 
 | |
8AWO
 
 | |
3JV7
 
 | | Structure of ADH-A from Rhodococcus ruber | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, ADH-A, ... | | Authors: | Karabec, M, Lyskowski, A, Gruber, K. | | Deposit date: | 2009-09-16 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into substrate specificity and solvent tolerance in alcohol dehydrogenase ADH-'A' from Rhodococcus ruber DSM 44541.
Chem.Commun.(Camb.), 2010
|
|
3GR8
 
 | | Structure of OYE from Geobacillus kaustophilus, orthorhombic crystal form | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase, ... | | Authors: | Uhl, M.K, Gruber, K. | | Deposit date: | 2009-03-25 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Old Yellow Enzyme-Catalyzed Dehydrogenation of Saturated Ketones
ADV.SYNTH.CATAL., 353, 2011
|
|
3GSY
 
 | | Structure of berberine bridge enzyme in complex with dehydroscoulerine | | Descriptor: | 2,9-dihydroxy-3,10-dimethoxy-5,6-dihydroisoquino[3,2-a]isoquinolinium, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Winkler, A, Macheroux, P, Gruber, K. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Berberine bridge enzyme catalyzes the six electron oxidation of (S)-reticuline to dehydroscoulerine.
Phytochemistry, 70, 2009
|
|
8A85
 
 | |
