4XHX
 
 | |
4XIK
 
 | |
4XJA
 
 | | Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 5-acetamido-2,3-difluoro-3-hydroxy-6-[1,2,3-trihydroxypropyl]oxane-2-carboxylic acid | | Descriptor: | (1s,3R,4S)-1-[(cyclohexylamino)methyl]-3,4-dihydroxycyclopentanesulfonic acid, (2R,3R,4R,5R,6R)-5-acetamido-2,3-difluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]tetrahydro-2H-pyran-2-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Brear, P. | | Deposit date: | 2015-01-08 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | `The Hunt for Serendipitous Allosteric Sites: Discovery of a novel allosteric inhibitor of the bacterial sialidase NanB
To be published
|
|
4XIL
 
 | |
4XMA
 
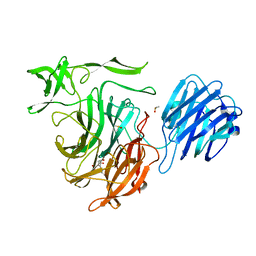 | |
2X0O
 
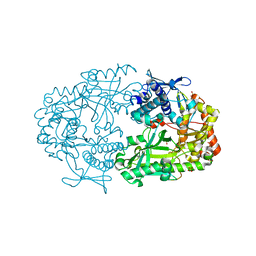 | | Apo structure of the Alcaligin biosynthesis protein C (AlcC) from Bordetella bronchiseptica | | Descriptor: | ALCALIGIN BIOSYNTHESIS PROTEIN, SULFATE ION | | Authors: | Johnson, K.A, Schmelz, S, Kadi, N, Mcmahon, S.A, Oke, M, Liu, H, Carter, L.G, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2009-12-16 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2VXZ
 
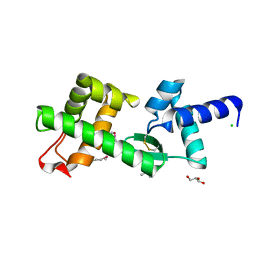 | | Crystal Structure of hypothetical protein PyrSV_gp04 from Pyrobaculum spherical virus | | Descriptor: | CHLORIDE ION, GLYCEROL, PYRSV_GP04 | | Authors: | Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Oke, M, Naismith, J.H, White, M.F. | | Deposit date: | 2008-07-15 | | Release date: | 2009-11-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2VW8
 
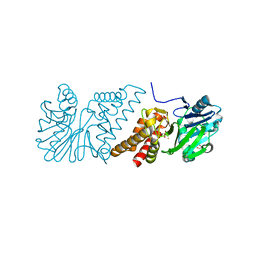 | | Crystal Structure of Quinolone signal response protein pqsE from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, FE (II) ION, ... | | Authors: | Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Oke, M, Naismith, J.H, White, M.F. | | Deposit date: | 2008-06-17 | | Release date: | 2010-07-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X5D
 
 | | Crystal Structure of a probable aminotransferase from Pseudomonas aeruginosa | | Descriptor: | PROBABLE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-08 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2WJ9
 
 | | ArdB | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Weikart, N.D, Roberts, G, Johnson, K.A, Oke, M, Cooper, L.P, McMahon, S.A, White, J.H, Liu, H, Carter, L.G, Walkinshaw, M.D, Blakely, G.W, Naismith, J.H, Dryden, D.T.F. | | Deposit date: | 2009-05-25 | | Release date: | 2010-08-18 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X3N
 
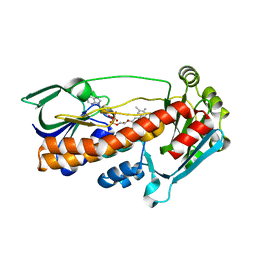 | | Crystal structure of pqsL, a probable FAD-dependent monooxygenase from Pseudomonas aeruginosa | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROBABLE FAD-DEPENDENT MONOOXYGENASE | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X4H
 
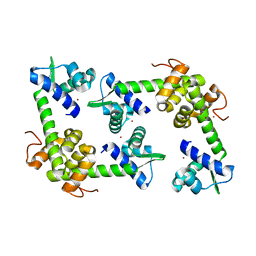 | | Crystal Structure of the hypothetical protein SSo2273 from Sulfolobus solfataricus | | Descriptor: | HYPOTHETICAL PROTEIN SSO2273, ZINC ION | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-31 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X4J
 
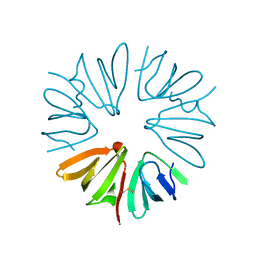 | | Crystal structure of ORF137 from Pyrobaculum spherical virus | | Descriptor: | HYPOTHETICAL PROTEIN ORF137 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-01 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X3E
 
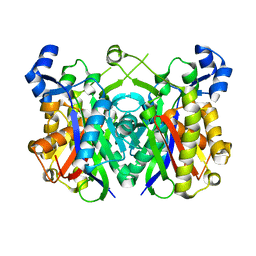 | | Crystal structure of 3-oxoacyl-(acyl carrier protein) synthase III, FabH from Pseudomonas aeruginosa PAO1 | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE 3 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X3L
 
 | | Crystal Structure of the Orn_Lys_Arg decarboxylase family protein SAR0482 from Methicillin-resistant Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, ORN/LYS/ARG DECARBOXYLASE FAMILY PROTEIN, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-21 | | Last modified: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X5C
 
 | | Crystal structure of hypothetical protein ORF131 from Pyrobaculum Spherical Virus | | Descriptor: | GLYCEROL, HYPOTHETICAL PROTEIN ORF131, ZINC ION | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-08 | | Release date: | 2010-07-21 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2X5Q
 
 | | Crystal Structure of Hypothetical protein sso1986 from Sulfolobus solfataricus P2 | | Descriptor: | SSO1986 | | Authors: | Oke, M, Carter, L, Johnson, K.A, Kerou, M, Liu, H, Mcmahon, S, Naismith, J.H, White, M.F. | | Deposit date: | 2010-02-10 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
1EUU
 
 | | SIALIDASE OR NEURAMINIDASE, LARGE 68KD FORM | | Descriptor: | SIALIDASE, SODIUM ION, beta-D-galactopyranose | | Authors: | Gaskell, A, Crennell, S.J, Taylor, G.L. | | Deposit date: | 1996-06-21 | | Release date: | 1997-01-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll.
Structure, 3, 1995
|
|
2X5P
 
 | | Crystal structure of the Streptococcus pyogenes fibronectin binding protein Fbab-B | | Descriptor: | FIBRONECTIN BINDING PROTEIN | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-10 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X4L
 
 | | Crystal structure of DesE, a ferric-siderophore receptor protein from Streptomyces coelicolor | | Descriptor: | FERRIC-SIDEROPHORE RECEPTOR PROTEIN | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X3G
 
 | | Crystal Structure of the hypothetical protein ORF119 from Sulfolobus islandicus rod-shaped virus 1 | | Descriptor: | SIRV1 HYPOTHETICAL PROTEIN ORF119 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-07-21 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2X3O
 
 | | Crystal Structure of the Hypothetical Protein PA0856 from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, GLYCEROL, HYPOTHETICAL PROTEIN PA0856 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X5G
 
 | | Crystal structure of the ORF131L51M mutant from Sulfolobus islandicus rudivirus 1 | | Descriptor: | CHLORIDE ION, MALONATE ION, ORF 131 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Naismith, J.H, White, M.F. | | Deposit date: | 2010-02-08 | | Release date: | 2010-07-21 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2X7I
 
 | | Crystal structure of mevalonate kinase from methicillin-resistant Staphylococcus aureus MRSA252 | | Descriptor: | CHLORIDE ION, CITRIC ACID, MEVALONATE KINASE | | Authors: | Oke, M, Yan, X, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-27 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
1EUS
 
 | | SIALIDASE COMPLEXED WITH 2-DEOXY-2,3-DEHYDRO-N-ACETYLNEURAMINIC ACID | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, SIALIDASE | | Authors: | Gaskell, A, Crennell, S.J, Taylor, G.L. | | Deposit date: | 1996-06-21 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll.
Structure, 3, 1995
|
|
