1YJT
 
 | | Solution structure of the Cu(I) form of the sixth soluble domain A69P mutant of Menkes protein | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S. | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
1YJR
 
 | | Solution structure of the apo form of the sixth soluble domain A69P mutant of Menkes protein | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
1YD6
 
 | | Crystal structure of the GIY-YIG N-terminal endonuclease domain of UvrC from Bacillus caldotenax | | Descriptor: | CHLORIDE ION, SULFATE ION, UvrC | | Authors: | Truglio, J.J, Rhau, B, Croteau, D.L, Wang, L, Skorvaga, M, Karakas, E, DellaVecchia, M.J, Wang, H, Van Houten, B, Kisker, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the first incision reaction during nucleotide excision repair
Embo J., 24, 2005
|
|
1YJV
 
 | | Solution structure of the Cu(I) form of the sixth soluble domain of Menkes protein | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S. | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
6CAA
 
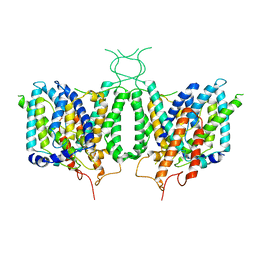 | | CryoEM structure of human SLC4A4 sodium-coupled acid-base transporter NBCe1 | | Descriptor: | Electrogenic sodium bicarbonate cotransporter 1 | | Authors: | Huynh, K.W, Jiang, J, Abuladze, N, Tsirulnikov, K, Kao, L, Shao, X, Newman, D, Azimov, R, Pushkin, A, Zhou, Z.H, Kurtz, I. | | Deposit date: | 2018-01-29 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | CryoEM structure of the human SLC4A4 sodium-coupled acid-base transporter NBCe1.
Nat Commun, 9, 2018
|
|
5DL9
 
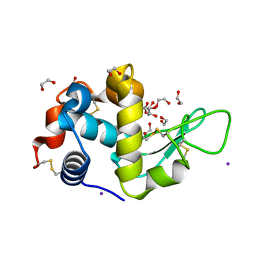 | | Structure of Tetragonal Lysozyme in complex with Iodine solved by UWO Students | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IODIDE ION, ... | | Authors: | Bednarski, R, Cirricione, N, Greco, A, Hodgson, R, Kent, S, McGowan, J, Notherm, B, Patt, M, Vue, L, Bianchetti, C.M. | | Deposit date: | 2015-09-04 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure of Tetragonal Lysozyme in complex with Iodine solved by UWO Students
To Be Published
|
|
6BRG
 
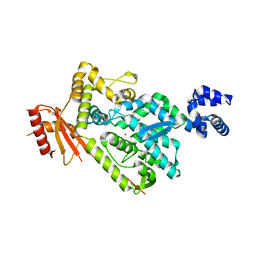 | | The SAM domain of mouse SAMHD1 is critical for its activation and regulation | | Descriptor: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, MAGNESIUM ION | | Authors: | Buzovetsky, O, Tang, C, Knecht, K.M, Antonucci, J.M, Wu, L, Ji, X, Xiong, Y. | | Deposit date: | 2017-11-30 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The SAM domain of mouse SAMHD1 is critical for its activation and regulation.
Nat Commun, 9, 2018
|
|
1Y8Z
 
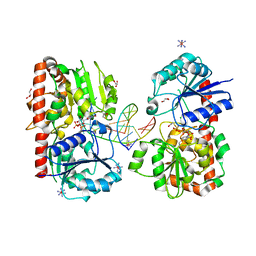 | | alpha-glucosyltransferase in complex with UDP and a 13-mer DNA containing a HMU base at 1.9 A resolution | | Descriptor: | 5'-D(*CP*TP*AP*TP*CP*TP*GP*AP*G)-3', 5'-D(*GP*AP*TP*AP*CP*TP*(5HU)P*AP*GP*AP*TP*AP*G)-3', CHLORIDE ION, ... | | Authors: | Lariviere, L, Sommer, N, Morera, S. | | Deposit date: | 2004-12-14 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural evidence of a passive base-flipping mechanism for AGT, an unusual GT-B glycosyltransferase.
J.Mol.Biol., 352, 2005
|
|
6BUV
 
 | | Structure of Mycobacterium tuberculosis NadD in complex with inhibitor [(1~{R},2~{R},5~{S})-5-methyl-2-propan-2-yl-cyclohexyl] 2-[3-methyl-2-(phenoxymethyl)benzimidazol-1-yl]ethanoate | | Descriptor: | 1-methyl-3-(2-{[(1R,2R,5S)-5-methyl-2-(propan-2-yl)cyclohexyl]oxy}-2-oxoethyl)-2-(phenoxymethyl)-1H-1,3-benzimidazol-3-ium, CHLORIDE ION, SODIUM ION, ... | | Authors: | Rodionova, I.A, Reed, R.W, Sorci, L, Osterman, A.L, Korotkov, K.V. | | Deposit date: | 2017-12-11 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Novel Antimycobacterial Compounds Suppress NAD Biogenesis by Targeting a Unique Pocket of NaMN Adenylyltransferase.
Acs Chem.Biol., 14, 2019
|
|
5DYF
 
 | | The crystal structure of Aminopeptidase N in complex with N-benzyl-1,2-diaminoethylphosphonic acid | | Descriptor: | Aminopeptidase N, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nocek, B, Joachimiak, A, Vassiliou, S, Berlicki, L, Mucha, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-09-24 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | The crystal structure of Aminopeptidase N in complex with N-benzyl-1,2-diaminoethylphosphonic acid
To Be Published
|
|
5DZ5
 
 | | Crystal structure of Dendroaspis polylepis mambalgin-1 wild-type in P41212 space group | | Descriptor: | Mambalgin-1 | | Authors: | Stura, E.A, Tepshi, L, Mourier, G, Kessler, P, Servent, D. | | Deposit date: | 2015-09-25 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mambalgin-1 Pain-relieving Peptide, Stepwise Solid-phase Synthesis, Crystal Structure, and Functional Domain for Acid-sensing Ion Channel 1a Inhibition.
J.Biol.Chem., 291, 2016
|
|
5DZS
 
 | | 1.5 Angstrom Crystal Structure of Shikimate Dehydrogenase 1 from Peptoclostridium difficile. | | Descriptor: | SULFATE ION, Shikimate dehydrogenase (NADP(+)) | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-26 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Shikimate Dehydrogenase 1 from Peptoclostridium difficile.
To Be Published
|
|
1YII
 
 | | Crystal Structures of Chicken Annexin V in Complex with Ca2+ | | Descriptor: | Annexin A5, CALCIUM ION, SULFATE ION | | Authors: | Ortlund, E.A, Chai, G, Genge, B, Wu, L.N.Y, Wuthier, R.E, Lebioda, L. | | Deposit date: | 2005-01-11 | | Release date: | 2005-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structures of Chicken Annexin A5 in Complex with Functional Modifiers Ca2+ and Zn2+ Reveal Zn2+ Induced Formation of Non-Planar Assemblies
Annexins, 1, 2005
|
|
5E1L
 
 | | Structural and functional analysis of the E. coli FtsZ interacting protein, ZapC, reveals insight into molecular properties of a novel Z ring stabilizing protein | | Descriptor: | Cell division protein ZapC | | Authors: | Schumacher, M.A, Huang, K.-H, Tchorzewski, L, Zeng, W, Janakiraman, A. | | Deposit date: | 2015-09-29 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Analyses Reveal Insights into the Molecular Properties of the Escherichia coli Z Ring Stabilizing Protein, ZapC.
J.Biol.Chem., 291, 2016
|
|
1XY5
 
 | | NMR strcutre of sst1-selective somatostatin (SRIF) analog 1 | | Descriptor: | SST1-selective somatosatin analog | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2011-09-28 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
1XYH
 
 | | Crystal Structure of Recombinant Human Cyclophilin J | | Descriptor: | cyclophilin-like protein PPIL3b | | Authors: | Huang, L.-L, Zhao, X.-M, Huang, C.-Q, Yu, L, Xia, Z.-X. | | Deposit date: | 2004-11-10 | | Release date: | 2005-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of recombinant human cyclophilin J, a novel member of the cyclophilin family.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6BZ0
 
 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD. | | Descriptor: | CHLORIDE ION, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-21 | | Release date: | 2018-01-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD.
To Be Published
|
|
6BZU
 
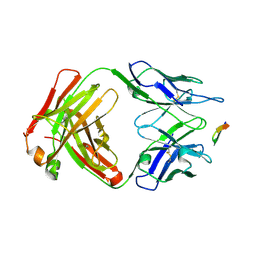 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the broadly neutralizing antibody 19B3 | | Descriptor: | 19B3 Heavy Chain, 19B3 Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Kong, L, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5E1J
 
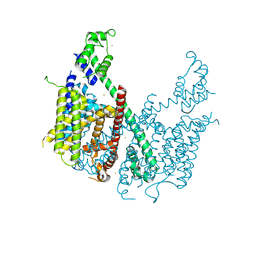 | | Structure of voltage-gated two-pore channel TPC1 from Arabidopsis thaliana | | Descriptor: | BARIUM ION, CALCIUM ION, Two pore calcium channel protein 1 | | Authors: | Guo, J, Zeng, W, Chen, Q, Lee, C, Chen, L, Yang, Y, Jiang, Y. | | Deposit date: | 2015-09-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.308 Å) | | Cite: | Structure of the voltage-gated two-pore channel TPC1 from Arabidopsis thaliana.
Nature, 531, 2016
|
|
5ETF
 
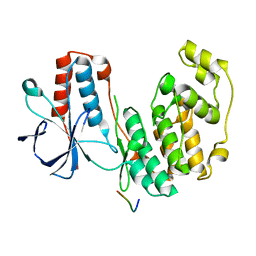 | | Structure of dead kinase MAPK14 with bound the KIM domain of MKK6 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 6, Mitogen-activated protein kinase 14 | | Authors: | Pellegrini, E, Palencia, A, Braun, L, Kapp, U, Bougdour, A, Belrhali, H, Bowler, M.W, Hakimi, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Subversion of MAP Kinase Signaling by an Intrinsically Disordered Parasite Secreted Agonist.
Structure, 25, 2017
|
|
1XL8
 
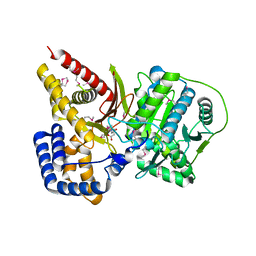 | | Crystal structure of mouse carnitine octanoyltransferase in complex with octanoylcarnitine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CARNITINE, OCTANOYLCARNITINE, ... | | Authors: | Jogl, G, Hsiao, Y.S, Tong, L. | | Deposit date: | 2004-09-30 | | Release date: | 2004-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of mouse carnitine octanoyltransferase and molecular determinants of substrate selectivity.
J.Biol.Chem., 280, 2005
|
|
6C1Q
 
 | | Crystal structure of human C5a receptor in complex with an orthosteric antagonist PMX53 and an allosteric antagonist NDT9513727 | | Descriptor: | 1-(1,3-benzodioxol-5-yl)-~{N}-(1,3-benzodioxol-5-ylmethyl)-~{N}-[(3-butyl-2,5-diphenyl-imidazol-4-yl)methyl]methanamine, PMX53, Soluble cytochrome b562, ... | | Authors: | Liu, H, Wang, L, Wei, Z, Zhang, C. | | Deposit date: | 2018-01-05 | | Release date: | 2018-05-30 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Orthosteric and allosteric action of the C5a receptor antagonists.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6BTA
 
 | | CypA Mutant - S99T C115S | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Fraser, J.S, Kenner, L.R, Liu, L. | | Deposit date: | 2017-12-06 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rescue of conformational dynamics in enzyme catalysis by directed evolution.
Nat Commun, 9, 2018
|
|
1XMC
 
 | | C323M mutant structure of mouse carnitine octanoyltransferase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Peroxisomal carnitine O-octanoyltransferase | | Authors: | Jogl, G, Hsiao, Y.S, Tong, L. | | Deposit date: | 2004-10-01 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mouse carnitine octanoyltransferase and molecular determinants of substrate selectivity.
J.Biol.Chem., 280, 2005
|
|
1XO5
 
 | | Crystal structure of CIB1, an EF-hand, integrin and kinase-binding protein | | Descriptor: | CALCIUM ION, Calcium and integrin-binding protein 1 | | Authors: | Gentry, H.R, Singer, A.U, Betts, L, Yang, C, Ferrara, J.D, Parise, L.V, Sondek, J. | | Deposit date: | 2004-10-05 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and Biochemical Characterization of CIB1 Delineates a New Family of EF-hand-containing Proteins
J.Biol.Chem., 280, 2005
|
|
