4S1P
 
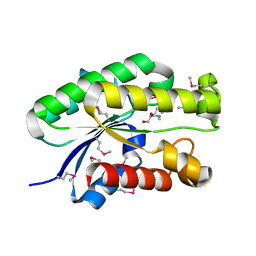 | | Shel_16390 protein, a putative SGNH hydrolase from Slackia heliotrinireducens | | Descriptor: | UNKNOWN LIGAND, Uncharacterized protein | | Authors: | Osipiuk, J, Cuff, M.E, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-14 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Shel_16390 protein, a putative SGNH hydrolase from Slackia heliotrinireducens
To be Published
|
|
4S1W
 
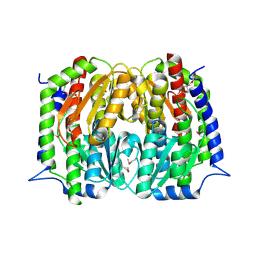 | | Structure of a putative Glutamine--Fructose-6-Phosphate Aminotransferase from Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] | | Authors: | Filippova, E.V, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-01-15 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of a putative Glutamine--Fructose-6-Phosphate Aminotransferase from Staphylococcus aureus subsp. aureus Mu50
To be Published
|
|
7RZP
 
 | | Crystal structure of putative NAD(P)H-flavin oxidoreductase from Haemophilus influenzae R2866 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Dihydropteridine reductase, ... | | Authors: | Maltseva, N, Kim, Y, Endres, M, Crofts, T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-08-27 | | Release date: | 2021-09-29 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional and Structural Characterization of Diverse NfsB Chloramphenicol Reductase Enzymes from Human Pathogens.
Microbiol Spectr, 10, 2022
|
|
4IXH
 
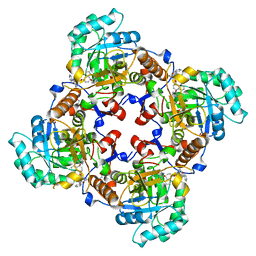 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Cryptosporidium parvum | | Descriptor: | (2S)-2-(naphthalen-1-yloxy)-N-[2-(pyridin-4-yl)-1,3-benzoxazol-5-yl]propanamide, 1,2-ETHANEDIOL, INOSINIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-25 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Optimization of Benzoxazole-Based Inhibitors of Cryptosporidium parvum Inosine 5'-Monophosphate Dehydrogenase.
J.Med.Chem., 56, 2013
|
|
4IYH
 
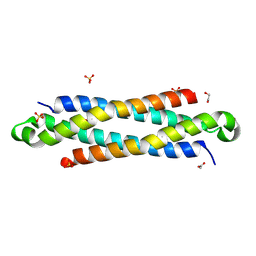 | | The crystal structure of a secreted protein EsxB (SeMet-labeled, C-term. His-Tagged) from Bacillus anthracis str. Sterne | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | The crystal structure of a secreted protein EsxB (SeMet-labeled, C-term. His-Tagged) from Bacillus anthracis str. Sterne
To be Published
|
|
7RZL
 
 | | Crystal structure of putative NAD(P)H-flavin oxidoreductase from Haemophilus influenzae R2846 in complex with 4-nitrophenol | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Maltseva, N, Kim, Y, Endres, M, Crofts, T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-08-27 | | Release date: | 2021-09-29 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Functional and Structural Characterization of Diverse NfsB Chloramphenicol Reductase Enzymes from Human Pathogens.
Microbiol Spectr, 10, 2022
|
|
7S14
 
 | | Crystal structure of putative NAD(P)H-flavin oxidoreductase from Haemophilus influenzae 86-028NP | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Crofts, T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-08-31 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Functional and Structural Characterization of Diverse NfsB Chloramphenicol Reductase Enzymes from Human Pathogens.
Microbiol Spectr, 10, 2022
|
|
7S1A
 
 | | Crystal structure of putative NAD(P)H-flavin oxidoreductase from Haemophilus influenzae Rd KW20 | | Descriptor: | ACETIC ACID, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Maltseva, N, Kim, Y, Endres, M, Crofts, T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-01 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Functional and Structural Characterization of Diverse NfsB Chloramphenicol Reductase Enzymes from Human Pathogens.
Microbiol Spectr, 10, 2022
|
|
4J42
 
 | | The crystal structure of a secreted protein EsxB (Mutant Y65F) from Bacillus anthracis str. Sterne | | Descriptor: | CITRATE ANION, FORMIC ACID, secreted protein EsxB | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-06 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The crystal structure of a secreted protein EsxB (Mutant Y65F) from Bacillus anthracis str. Sterne
To be Published
|
|
4IT5
 
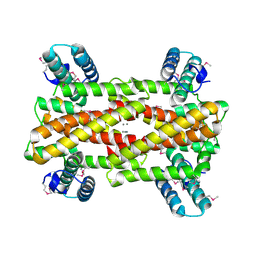 | | Chaperone HscB from Vibrio cholerae | | Descriptor: | CALCIUM ION, Co-chaperone protein HscB homolog | | Authors: | Osipiuk, J, Gu, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-17 | | Release date: | 2013-01-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Chaperone HscB from Vibrio cholerae.
To be Published
|
|
4RT5
 
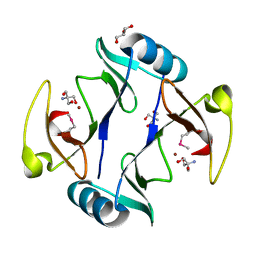 | | The crystal structure of a glyoxalase/bleomycin resistance protein/dioxygenase protein from planctomyces limnophilus dsm 3776 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Glyoxalase/bleomycin resistance protein/dioxygenase, ... | | Authors: | Wu, R, Bearden, J, Kim, Y, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-13 | | Release date: | 2014-12-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a glyoxalase/bleomycin resistance protein/dioxygenase protein from Planctomyces limnophilus dsm 3776
TO BE PUBLISHED
|
|
4J7J
 
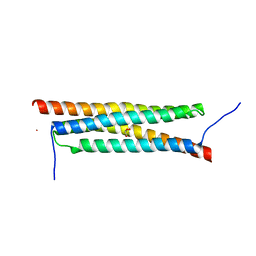 | | The crystal structure of a secreted protein EsxB (Mutant G53A) from Bacillus anthracis str. Sterne | | Descriptor: | GLYCEROL, secreted protein EsxB | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-13 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.458 Å) | | Cite: | The crystal structure of a secreted protein EsxB (Mutant, G53A) from Bacillus anthracis str. Sterne
To be Published
|
|
4RU0
 
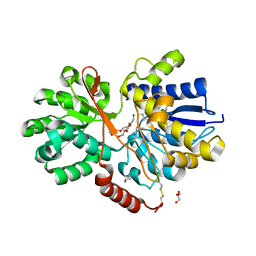 | | The crystal structure of abc transporter permease from pseudomonas fluorescens group | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECANE, GLYCEROL, Putative branched-chain amino acid ABC transporter, ... | | Authors: | Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-17 | | Release date: | 2014-11-26 | | Method: | X-RAY DIFFRACTION (2.442 Å) | | Cite: | The crystal structure of abc transporter permease from pseudomonas fluorescens group
To be Published
|
|
4RYK
 
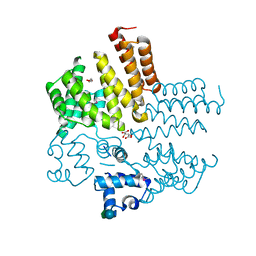 | | Crystal structure of a putative transcriptional regulator from Listeria monocytogenes EGD-e | | Descriptor: | DI(HYDROXYETHYL)ETHER, L(+)-TARTARIC ACID, Lmo0325 protein, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-15 | | Release date: | 2015-01-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a putative transcriptional regulator from Listeria monocytogenes EGD-e
To be Published
|
|
4RM1
 
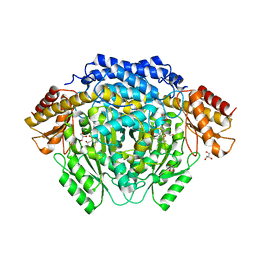 | |
7KOJ
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder494 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]-5-{[(prop-2-en-1-yl)carbamoyl]amino}benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder494
to be published
|
|
4RTF
 
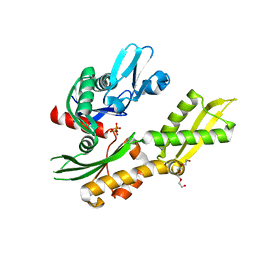 | | Crystal structure of molecular chaperone DnaK from Mycobacterium tuberculosis H37Rv | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK, TETRAETHYLENE GLYCOL | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Endres, M, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2014-11-14 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal structure of molecular chaperone DnaK from Mycobacterium tuberculosis H37Rv
To be Published
|
|
4RUW
 
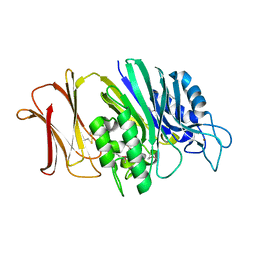 | |
4JGQ
 
 | | The crystal structure of sporulation kinase D mutant sensor domain, r131a, from Bacillus subtilis subsp in co-crystallization with pyruvate | | Descriptor: | ACETIC ACID, Sporulation kinase D | | Authors: | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
7KYU
 
 | | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1H-indole-5-carbonyl)oxy]-1H-benzotriazole, 3C-like proteinase | | Authors: | Tan, K, Maltseva, N.I, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate
To Be Published
|
|
4JJT
 
 | | The crystal structure of enoyl-CoA hydratase from Mycobacterium tuberculosis H37Rv | | Descriptor: | ACETATE ION, Enoyl-CoA hydratase, GLYCEROL | | Authors: | Tan, K, Holowicki, J, Endres, M, Kim, C.-Y, Kim, H, Hung, L.-W, Terwilliger, T.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-03-08 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | The crystal structure of enoyl-CoA hydratase from Mycobacterium tuberculosis H37Rv
To be Published
|
|
7KFF
 
 | | Crystal structure of TrmD tRNA (guanine-N1)-methyltransferase from Corynebacterium diphtheriae in complex with SAH | | Descriptor: | ACETATE ION, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Michalska, K, Tanase, L, Maltseva, N, Kim, Y, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-10-13 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of TrmD tRNA (guanine-N1)-methyltransferase from Corynebacterium diphtheriae in complex with SAH
To Be Published
|
|
4J7K
 
 | | The crystal structure of a secreted protein EsxB (Mutant E54Q) from Bacillus anthracis str. Sterne | | Descriptor: | secreted protein EsxB | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-13 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The crystal structure of a secreted protein EsxB (Mutant E54Q) from Bacillus anthracis str. Sterne
To be Published
|
|
7KOK
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder496 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder496
to be published
|
|
4O2I
 
 | | The crystal structure of non-LEE encoded type III effector C from Citrobacter rodentium | | Descriptor: | Non-LEE encoded type III effector C, ZINC ION | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Adkins, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of non-LEE encoded type III effector C from Citrobacter rodentium
To be Published
|
|
