3X42
 
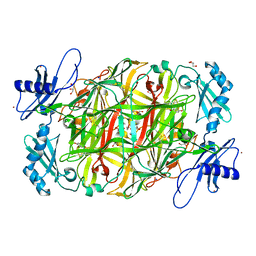 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis in the presence of sodium bromide | | Descriptor: | BROMIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.875 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X41
 
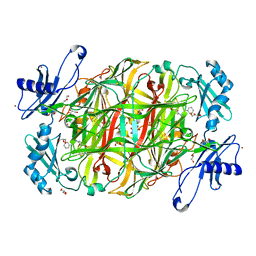 | | Copper amine oxidase from Arthrobacter globiformis: Product Schiff-base form produced by anaerobic reduction in the presence of sodium bromide | | Descriptor: | BROMIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X23
 
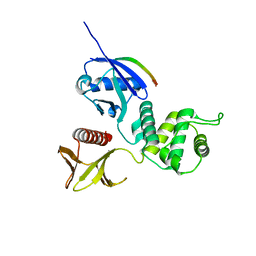 | | Radixin complex | | Descriptor: | Peptide from Matrix metalloproteinase-14, Radixin | | Authors: | Terawaki, S, Kitano, K, Aoyama, M, Mori, T, Hakoshima, T. | | Deposit date: | 2014-12-09 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | MT1-MMP recognition by ERM proteins and its implication in CD44 shedding
Genes Cells, 20, 2015
|
|
3X3X
 
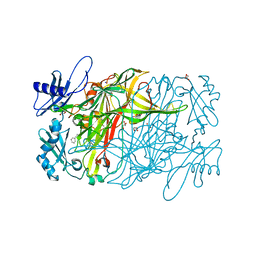 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine | | Descriptor: | 2-PHENYL-ETHANOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X3Z
 
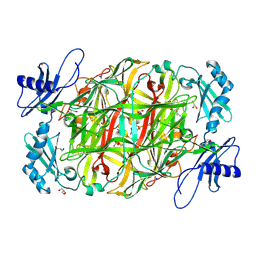 | | Copper amine oxidase from Arthrobacter globiformis: Aminoresorcinol form produced by anaerobic reduction with ethylamine hydrochloride | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3UKJ
 
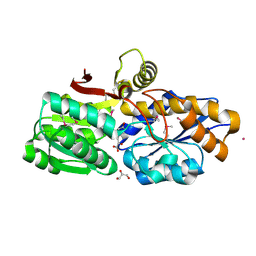 | | Crystal structure of extracellular ligand-binding receptor from Rhodopseudomonas palustris HaA2 | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, Extracellular ligand-binding receptor, GLYCEROL, ... | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-11-09 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
3UK0
 
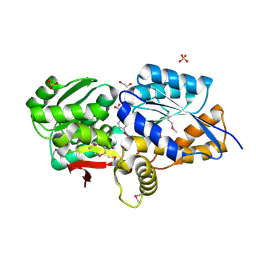 | | RPD_1889 protein, an extracellular ligand-binding receptor from Rhodopseudomonas palustris. | | Descriptor: | 1,2-ETHANEDIOL, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, Extracellular ligand-binding receptor, ... | | Authors: | Osipiuk, J, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-11-08 | | Release date: | 2011-11-23 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
3OON
 
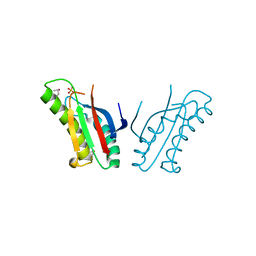 | | The structure of an outer membrance protein from Borrelia burgdorferi B31 | | Descriptor: | Outer membrane protein (Tpn50), SULFATE ION | | Authors: | Fan, Y, Bigelow, L, Feldman, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Insights into PG-binding, conformational change, and dimerization of the OmpA C-terminal domains from Salmonella enterica serovar Typhimurium and Borrelia burgdorferi.
Protein Sci., 26, 2017
|
|
4RHA
 
 | | Structure of the C-terminal domain of outer-membrane protein OmpA from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Outer membrane protein A, ... | | Authors: | Cuff, M.E, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-01 | | Release date: | 2014-10-29 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into PG-binding, conformational change, and dimerization of the OmpA C-terminal domains from Salmonella enterica serovar Typhimurium and Borrelia burgdorferi.
Protein Sci., 26, 2017
|
|
3TX6
 
 | | The Structure of a putative ABC-transporter periplasmic component from Rhodopseudomonas palustris | | Descriptor: | 1,2-ETHANEDIOL, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, ACETIC ACID, ... | | Authors: | Cuff, M.E, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-22 | | Release date: | 2011-11-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4EYQ
 
 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris HaA2 in complex with caffeic acid/3-(4-HYDROXY-PHENYL)PYRUVIC ACID | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, CAFFEIC ACID, Extracellular ligand-binding receptor | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4F8J
 
 | | The structure of an aromatic compound transport protein from Rhodopseudomonas palustris in complex with p-coumarate | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Cuff, M.E, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-17 | | Release date: | 2012-09-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4EYO
 
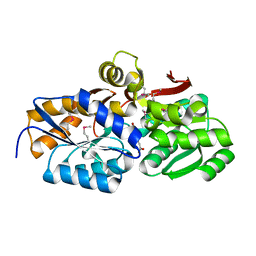 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris HaA2 in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Extracellular ligand-binding receptor | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4FB4
 
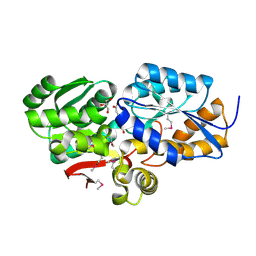 | | The Structure of an ABC-Transporter Family Protein from Rhodopseudomonas palustris in Complex with Caffeic Acid | | Descriptor: | CAFFEIC ACID, GLYCEROL, Putative branched-chain amino acid transport system substrate-binding protein | | Authors: | Cuff, M.E, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-22 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
2BI6
 
 | | NMR STUDY OF BROMELAIN INHIBITOR VI FROM PINEAPPLE STEM | | Descriptor: | BROMELAIN INHIBITOR VI | | Authors: | Hatano, K.-I. | | Deposit date: | 1995-12-07 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of bromelain inhibitor IV from pineapple stem: structural similarity with Bowman-Birk trypsin/chymotrypsin inhibitor from soybean.
Biochemistry, 35, 1996
|
|
1BI6
 
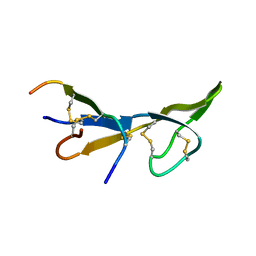 | | NMR STRUCTURE OF BROMELAIN INHIBITOR VI FROM PINEAPPLE STEM | | Descriptor: | BROMELAIN INHIBITOR VI | | Authors: | Hatano, K.-I. | | Deposit date: | 1995-12-07 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of bromelain inhibitor IV from pineapple stem: structural similarity with Bowman-Birk trypsin/chymotrypsin inhibitor from soybean.
Biochemistry, 35, 1996
|
|
5GWF
 
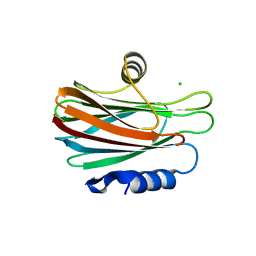 | | FraC with GlcNAc(6S) bound | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, CHLORIDE ION, DELTA-actitoxin-Afr1a, ... | | Authors: | Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2016-09-11 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Haemolytic actinoporins interact with carbohydrates using their lipid-binding module
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 372, 2017
|
|
5SZD
 
 | | Crystal structure of Aquifex aeolicus Hfq at 1.5A | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Stanek, K, Patterson, J, Randolph, P.S, Mura, C. | | Deposit date: | 2016-08-13 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.494 Å) | | Cite: | Crystal structure and RNA-binding properties of an Hfq homolog from the deep-branching Aquificae: conservation of the lateral RNA-binding mode.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5SZE
 
 | | Crystal structure of Aquifex aeolicus Hfq-RNA complex at 1.5A | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, DI(HYDROXYETHYL)ETHER, RNA (5'-R(P*UP*UP*U)-3'), ... | | Authors: | Stanek, K, Patterson, J, Randolph, P.S, Mura, C. | | Deposit date: | 2016-08-13 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure and RNA-binding properties of an Hfq homolog from the deep-branching Aquificae: conservation of the lateral RNA-binding mode.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
2DB4
 
 | | Crystal structure of rotor ring with DCCD of the V- ATPase from Enterococcus hirae | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, DICYCLOHEXYLUREA, SODIUM ION, ... | | Authors: | Murata, T, Yamato, I, Kakinuma, Y, Shirouzu, M, Walker, J.E, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the rotor ring modified with N,N'-dicyclohexylcarbodiimide of the Na+-transporting vacuolar ATPase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4EJF
 
 | |
6PU9
 
 | | Crystal Structure of the Type B Chloramphenicol O-Acetyltransferase from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase, CHLORIDE ION | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Grimshaw, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-17 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of three Type B and C chloramphenicol acetyltransferases from Vibrio species.
Protein Sci., 29, 2020
|
|
6PUA
 
 | | The 2.0 A Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Stam, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional characterization of three Type B and C chloramphenicol acetyltransferases from Vibrio species.
Protein Sci., 29, 2020
|
|
6WN5
 
 | | 1.52 Angstrom Resolution Crystal Structure of Transcriptional Regulator HdfR from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, Transcriptional regulator HdfR | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6WN8
 
 | | 2.70 Angstrom Resolution Crystal Structure of Uracil Phosphoribosyl Transferase from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, SULFATE ION, Uracil phosphoribosyltransferase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
