7XMA
 
 | | Crystal structure of Bovine heart cytochrome c oxidase, apo structure with DMSO | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7XMB
 
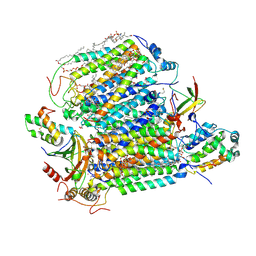 | | Crystal structure of Bovine heart cytochrome c oxidase, the structure complexed with an allosteric inhibitor T113 | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Shintani, Y, Takashima, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
4M09
 
 | | Crystal Structure of Mutant Chlorite Dismutase from Candidatus Nitrospira defluvii W146Y R173Q | | Descriptor: | 1,2-ETHANEDIOL, Chlorite dismutase, IMIDAZOLE, ... | | Authors: | Gysel, K, Hagmueller, A, Djinovic-Carugo, K. | | Deposit date: | 2013-08-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Manipulating conserved heme cavity residues of chlorite dismutase: effect on structure, redox chemistry, and reactivity.
Biochemistry, 53, 2014
|
|
2GRX
 
 | | Crystal structure of TonB in complex with FhuA, E. coli outer membrane receptor for ferrichrome | | Descriptor: | 2-AMINO-VINYL-PHOSPHATE, 3-HYDROXY-TETRADECANOIC ACID, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-[L-glycero-alpha-D-manno-heptopyranose-(1-5)]3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2,3-dideoxy-alpha-D-glucoyranose-(1-6)-2-amino-2,3-dideoxy-alpha-D-glucoyranose, ... | | Authors: | Pawelek, P.D, Allaire, M, Coulton, J.W. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of TonB in complex with FhuA, E. coli outer membrane receptor.
Science, 312, 2006
|
|
2POX
 
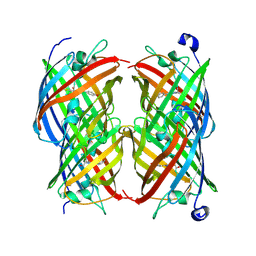 | |
3C41
 
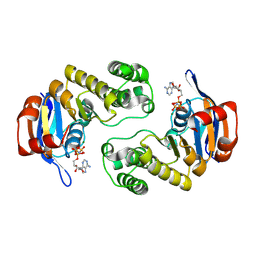 | | ABC protein ArtP in complex with AMP-PNP/Mg2+ | | Descriptor: | Amino acid ABC transporter (ArtP), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Thaben, P.F, Eckey, V, Scheffel, F, Saenger, W, Schneider, E, Vahedi-Faridi, A. | | Deposit date: | 2008-01-29 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures of the ATP-binding cassette (ABC) Protein ArtP from Geobacillus stearothermophilus in complexes with nucleotides and nucleotide analogs reveal an intermediate semiclosed dimer in the post hydrolyses state and an asymmetry in the dimerisation region
To be Published
|
|
2Z9A
 
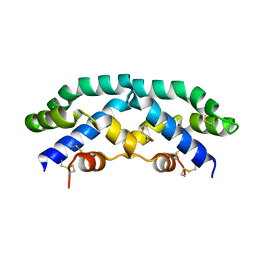 | |
3C4J
 
 | | ABC protein ArtP in complex with ATP-gamma-S | | Descriptor: | Amino acid ABC transporter (ArtP), MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Thaben, P.F, Eckey, V, Scheffel, F, Saenger, W, Schneider, E, Vahedi-Faridi, A. | | Deposit date: | 2008-01-30 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structures of the ATP-binding cassette (ABC) Protein ArtP from Geobacillus stearothermophilus in complexes with nucleotides and nucleotide analogs reveal an intermediate semiclosed dimer in the post hydrolyses state and an asymmetry in the dimerisation region
To be Published
|
|
2Q2C
 
 | | Crystal structures of the arginine-, lysine-, histidine-binding protein ArtJ from the thermophilic bacterium Geobacillus stearothermophilus | | Descriptor: | ArtJ, GLYCEROL, HISTIDINE, ... | | Authors: | Vahedi-Faridi, A, Scheffel, F, Eckey, V, Saenger, W, Schneider, E. | | Deposit date: | 2007-05-28 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures and mutational analysis of the arginine-, lysine-, histidine-binding protein ArtJ from Geobacillus stearothermophilus. Implications for interactions of ArtJ with its cognate ATP-binding cassette transporter, Art(MP)2
J.Mol.Biol., 375, 2008
|
|
2PVU
 
 | | Crystal structures of the arginine-, lysine-, histidine-binding protein ArtJ from the thermophilic bacterium Geobacillus stearothermophilus | | Descriptor: | ArtJ, LYSINE, SULFATE ION | | Authors: | Vahedi-Faridi, A, Scheffel, F, Eckey, V, Saenger, W, Schneider, E. | | Deposit date: | 2007-05-10 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures and mutational analysis of the arginine-, lysine-, histidine-binding protein ArtJ from Geobacillus stearothermophilus. Implications for interactions of ArtJ with its cognate ATP-binding cassette transporter, Art(MP)2
J.Mol.Biol., 375, 2008
|
|
2LEV
 
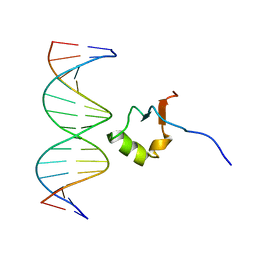 | | Structure of the DNA complex of the C-Terminal domain of Ler | | Descriptor: | DNA (5'-D(*CP*CP*TP*AP*TP*CP*AP*AP*TP*TP*AP*TP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*AP*TP*AP*AP*TP*TP*GP*AP*TP*AP*GP*G)-3'), Ler | | Authors: | Cordeiro, T.N, Schimdt, H, Madrid, C, Juarez, A, Bernado, P, Grisienger, C, Garcia, J, Pons, M. | | Deposit date: | 2011-06-23 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Indirect DNA Readout by an H-NS Related Protein: Structure of the DNA Complex of the C-Terminal Domain of Ler.
Plos Pathog., 7, 2011
|
|
2QYP
 
 | |
7ZBN
 
 | |
1AUW
 
 | | H91N DELTA 2 CRYSTALLIN FROM DUCK | | Descriptor: | DELTA 2 CRYSTALLIN | | Authors: | Abu-Abed, M, Vallee, F, Howell, P.L. | | Deposit date: | 1997-09-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural comparison of the enzymatically active and inactive forms of delta crystallin and the role of histidine 91.
Biochemistry, 36, 1997
|
|
3CJ5
 
 | |
3CUI
 
 | |
2R0R
 
 | |
1VIO
 
 | | Crystal structure of pseudouridylate synthase | | Descriptor: | 1,4-BUTANEDIOL, Ribosomal small subunit pseudouridine synthase A | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure of the pseudouridine synthase RsuA from Haemophilus influenzae.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
4M06
 
 | | Crystal Structure of Mutant Chlorite Dismutase from Candidatus Nitrospira defluvii W145F in Complex with Cyanide | | Descriptor: | 1,2-ETHANEDIOL, CYANIDE ION, Chlorite dismutase, ... | | Authors: | Hagmueller, A, Gysel, K, Djinovic-Carugo, K. | | Deposit date: | 2013-08-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Manipulating conserved heme cavity residues of chlorite dismutase: effect on structure, redox chemistry, and reactivity.
Biochemistry, 53, 2014
|
|
3CIZ
 
 | |
1VJE
 
 | |
3CUH
 
 | |
3CUG
 
 | |
1VHN
 
 | |
1VI7
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yigZ | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of YIGZ, a conserved hypothetical protein from Escherichia coli k12 with a novel fold
Proteins, 55, 2004
|
|
