3TO3
 
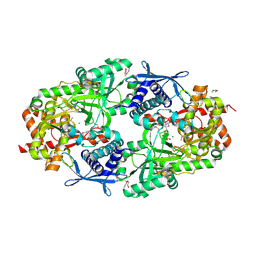 | | Crystal Structure of Petrobactin Biosynthesis Protein AsbB from Bacillus anthracis str. Sterne | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Kim, Y, Eschenfeldt, W, Stols, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-03 | | Release date: | 2011-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Functional and Structural Analysis of the Siderophore Synthetase AsbB through Reconstitution of the Petrobactin Biosynthetic Pathway from Bacillus anthracis.
J.Biol.Chem., 287, 2012
|
|
7KB9
 
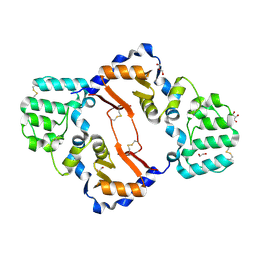 | | THE STRUCTURE OF A SENSOR DOMAIN OF A HISTIDINE KINASE (VxrA) FROM VIBRIO CHOLERAE O1 BIOVAR ELTOR STR. N16961, D238-T240 deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Sensor histidine kinase | | Authors: | Tan, K, Wu, R, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 2021
|
|
7KOL
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496
to be published
|
|
1NI9
 
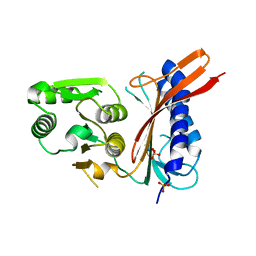 | | 2.0 A structure of glycerol metabolism protein from E. coli | | Descriptor: | Protein glpX, SULFATE ION | | Authors: | Sanishvili, R, Brunzelle, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-23 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
3TZL
 
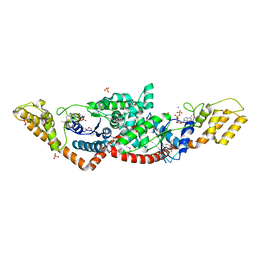 | | Crystal Structure of Tryptophanyl-tRNA Synthetase from Campylobacter jejuni complexed with ADP and Tryptophane | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Kim, Y, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-27 | | Release date: | 2011-10-05 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Crystal Structure of Tryptophanyl-tRNA Synthetase from Campylobacter jejuni complexed with ADP and Tryptophane
To be Published
|
|
3TP9
 
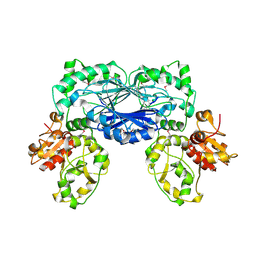 | | Crystal structure of Alicyclobacillus acidocaldarius protein with beta-lactamase and rhodanese domains | | Descriptor: | BETA-LACTAMASE and RHODANESE DOMAIN PROTEIN, ZINC ION | | Authors: | Michalska, K, Chhor, G, Mandel, M.E, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-07 | | Release date: | 2011-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Alicyclobacillus acidocaldarius protein with beta-lactamase and rhodanese domains
To be Published
|
|
7KFF
 
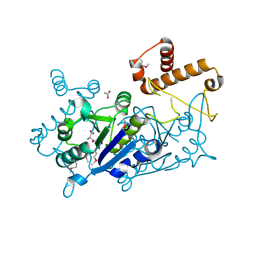 | | Crystal structure of TrmD tRNA (guanine-N1)-methyltransferase from Corynebacterium diphtheriae in complex with SAH | | Descriptor: | ACETATE ION, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Michalska, K, Tanase, L, Maltseva, N, Kim, Y, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-10-13 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of TrmD tRNA (guanine-N1)-methyltransferase from Corynebacterium diphtheriae in complex with SAH
To Be Published
|
|
7KOK
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder496 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder496
to be published
|
|
3TVA
 
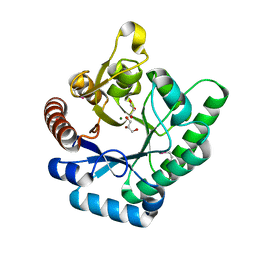 | | Crystal Structure of Xylose isomerase domain protein from Planctomyces limnophilus | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, Y, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-19 | | Release date: | 2011-10-05 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Crystal Structure of Xylose isomerase domain protein from Planctomyces limnophilus
To be Published
|
|
3U2E
 
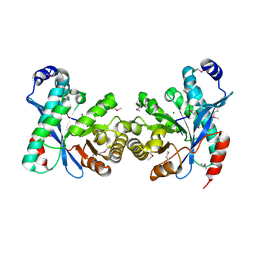 | | EAL domain of phosphodiesterase PdeA in complex with 5'-pGpG and Mg++ | | Descriptor: | GGDEF family protein, MAGNESIUM ION, RNA (5'-R(P*GP*G)-3') | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Massa, C, Schirmer, T, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-10-03 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | EAL domain from Caulobacter crescentus in complex with 5'-pGpG and Mg++
To be Published
|
|
1NOG
 
 | | Crystal Structure of Conserved Protein 0546 from Thermoplasma Acidophilum | | Descriptor: | conserved hypothetical protein TA0546 | | Authors: | Saridakis, V, Sanishvili, R, Iakounine, A, Xu, X, Pennycooke, M, Gu, J, Joachimiak, A, Arrowsmith, C.H, Edwards, A.M, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-16 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structural basis for methylmalonic aciduria. The crystal structure of archaeal ATP:cobalamin adenosyltransferase.
J.Biol.Chem., 279, 2004
|
|
3UFG
 
 | | The crystal structure of glycyl-tRNA synthetase subunit alpha from Campylobacter jejuni subsp. jejuni NCTC in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Glycyl-tRNA synthetase alpha subunit, LEUCINE | | Authors: | Tan, K, Zhou, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-01 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | The crystal structure of glycyl-tRNA synthetase subunit alpha from Campylobacter jejuni subsp. jejuni NCTC in complex with ATP
To be Published
|
|
4YCS
 
 | | Crystal structure of putative lipoprotein from Peptoclostridium difficile 630 (fragment) | | Descriptor: | ACETATE ION, GLYCEROL, SODIUM ION, ... | | Authors: | Michalska, K, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-20 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of putative lipoprotein from Peptoclostridium difficile 630 (fragment)
To Be Published
|
|
4YF1
 
 | | 1.85 angstrom crystal structure of lmo0812 from Listeria monocytogenes EGD-e | | Descriptor: | CITRATE ANION, Lmo0812 protein, SODIUM ION | | Authors: | Krishna, S.N, Light, S.H, Filippova, E.V, Minasov, G, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 angstrom crystal structure of lmo0812 from Listeria monocytogenes EGD-e
To Be Published
|
|
3FH3
 
 | | Crystal structure of a putative ECF-type sigma factor negative effector from Bacillus anthracis str. Sterne | | Descriptor: | NICKEL (II) ION, putative ECF-type sigma factor negative effector | | Authors: | Nocek, B, Kim, Y, Joachimiak, G, Du, J, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-08 | | Release date: | 2009-01-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal structure of a putative ECF-type sigma factor negative effector from Bacillus anthracis str. Sterne
To be Published
|
|
4Y7D
 
 | | Alpha/beta hydrolase fold protein from Nakamurella multipartita | | Descriptor: | Alpha/beta hydrolase fold protein, CHLORIDE ION, SODIUM ION | | Authors: | Cuff, M.E, OSIPIUK, J, Holowicki, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-14 | | Release date: | 2015-02-25 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Alpha/beta hydrolase fold protein from Nakamurella multipartita.
to be published
|
|
3UF6
 
 | | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoD (3'-dephosphocoenzyme A) | | Descriptor: | DEPHOSPHO COENZYME A, Lmo1369 protein | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoD (3'-dephosphocoenzyme A)
To be Published
|
|
3UHF
 
 | | Crystal Structure of Glutamate Racemase from Campylobacter jejuni subsp. jejuni | | Descriptor: | CHLORIDE ION, D-GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Maltseva, N, Mulligan, R, Kwon, K, Kim, Y, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-03 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Glutamate Racemase
from Campylobacter jejuni subsp. jejuni
To be Published
|
|
4E5S
 
 | |
3UXV
 
 | | Crystal Structure of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with NADP and PreQ | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, GUANINE, ... | | Authors: | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-05 | | Release date: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal Structure of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with NADP and PreQ
To be Published, 2012
|
|
3UO2
 
 | | Jac1 co-chaperone from Saccharomyces cerevisiae | | Descriptor: | J-type co-chaperone JAC1, mitochondrial | | Authors: | Osipiuk, J, Mulligan, R, Bigelow, L, Marszalek, J, Craig, E.A, Dutkiewicz, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Interaction of j-protein co-chaperone jac1 with fe-s scaffold isu is indispensable in vivo and conserved in evolution.
J.Mol.Biol., 417, 2012
|
|
5BMO
 
 | | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus | | Descriptor: | ACETATE ION, POTASSIUM ION, Putative uncharacterized protein LnmX | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Cuff, M, Endres, M, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus.
to be published
|
|
1TD5
 
 | | Crystal Structure of the Ligand Binding Domain of E. coli IclR. | | Descriptor: | Acetate operon repressor | | Authors: | Walker, J.R, Evdokimova, L, Zhang, R.-G, Bochkarev, A, Joachimiak, A, Arrowsmith, C, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-21 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analyses of the Ligand Binding Sites of the IclR family of transcriptional regulators
To be Published
|
|
3TEV
 
 | | The crystal structure of glycosyl hydrolase from Deinococcus radiodurans R1 | | Descriptor: | Glycosyl hyrolase, family 3 | | Authors: | Chang, C, Hatzos-Skintges, C, Kohler, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-08-15 | | Release date: | 2011-08-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of glycosyl hydrolase from Deinococcus radiodurans R1
To be Published
|
|
3TNG
 
 | | The crystal structure of a possible phosphate acetyl/butaryl transferase from Listeria monocytogenes EGD-e. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lmo1369 protein, NICKEL (II) ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-01 | | Release date: | 2011-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The crystal structure of a possible phosphate acetyl/butaryl transferase from Listeria monocytogenes EGD-e.
To be Published
|
|
