5ZP7
 
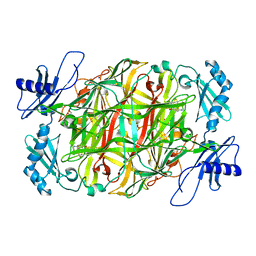 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 6 at 277 K (3) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPM
 
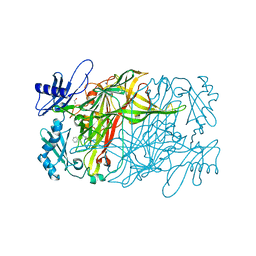 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine at pH 7 at 288 K (2) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4P79
 
 | | Crystal structure of mouse claudin-15 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Claudin-15 | | Authors: | Suzuki, H, Nishizawa, T, Tani, K, Yamazaki, Y, Tamura, A, Ishitani, R, Dohmae, N, Tsukita, S, Nureki, O, Fujiyoshi, Y. | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a claudin provides insight into the architecture of tight junctions.
Science, 344, 2014
|
|
6AGP
 
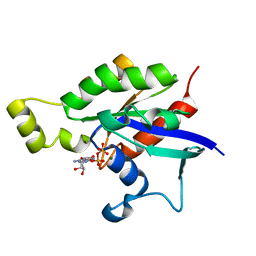 | | Structure of Rac1 in the low-affinity state for Mg2+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Toyama, Y, Kontani, K, Katada, T, Shimada, I. | | Deposit date: | 2018-08-13 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational landscape alternations promote oncogenic activities of Ras-related C3 botulinum toxin substrate 1 as revealed by NMR.
Sci Adv, 5, 2019
|
|
1G3B
 
 | | BOVINE BETA-TRYPSIN BOUND TO META-AMIDINO SCHIFF BASE MAGNESIUM(II) CHELATE | | Descriptor: | 2-(5-CARBAMIMIDOYL-2-HYDROXY-BENZYLAMINO)-PROPIONIC ACID, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Toyota, E, Ng, K.K.S, Sekizaki, H, Itoh, K, Tanizawa, K, James, M.N.G. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic analyses of complexes between bovine beta-trypsin and Schiff base copper(II) or iron(III) chelates.
J.Mol.Biol., 305, 2001
|
|
1G3D
 
 | | BOVINE BETA-TRYPSIN BOUND TO META-AMIDINO SCHIFF BASE COPPER (II) CHELATE | | Descriptor: | 2-(5-CARBAMIMIDOYL-2-HYDROXY-BENZYLAMINO)-PROPIONIC ACID, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Toyota, E, Ng, K.K.S, Sekizaki, H, Itoh, K, Tanizawa, K, James, M.N.G. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic analyses of complexes between bovine beta-trypsin and Schiff base copper(II) or iron(III) chelates.
J.Mol.Biol., 305, 2001
|
|
1G3C
 
 | | BOVINE BETA-TRYPSIN BOUND TO PARA-AMIDINO SCHIFF BASE IRON(III) CHELATE | | Descriptor: | 2-(4-CARBAMIMIDOYL-2-HYDROXY-BENZYLAMINO)-PROPIONIC ACID, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Toyota, E, Ng, K.K.S, Sekizaki, H, Itoh, K, Tanizawa, K, James, M.N.G. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic analyses of complexes between bovine beta-trypsin and Schiff base copper(II) or iron(III) chelates.
J.Mol.Biol., 305, 2001
|
|
1G3E
 
 | | BOVINE BETA-TRYPSIN BOUND TO PARA-AMIDINO SCHIFF-BASE COPPER (II) CHELATE | | Descriptor: | 2-(4-CARBAMIMIDOYL-2-HYDROXY-BENZYLAMINO)-PROPIONIC ACID, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Toyota, E, Ng, K.K.S, Sekizaki, H, Itoh, K, Tanizawa, K, James, M.N.G. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic analyses of complexes between bovine beta-trypsin and Schiff base copper(II) or iron(III) chelates.
J.Mol.Biol., 305, 2001
|
|
2Z84
 
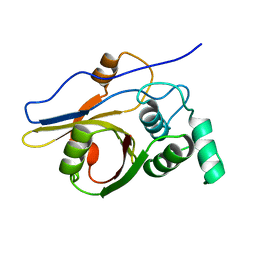 | | Insights from crystal and solution structures of mouse UfSP1 | | Descriptor: | Ufm1-specific protease 1 | | Authors: | Ha, B.H, Ahn, H.C, Kang, S.H, Tanaka, K, Chung, C.H, Kim, E.E. | | Deposit date: | 2007-08-30 | | Release date: | 2008-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for Ufm1 processing by UfSP1
J. Biol. Chem., 283, 2008
|
|
6KKA
 
 | | Xylanase J mutant from Bacillus sp. 41M-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Suzuki, M, Takita, T, Nakatani, K. | | Deposit date: | 2019-07-24 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Increase in the thermostability of GH11 xylanase XynJ from Bacillus sp. strain 41M-1 using site saturation mutagenesis.
Enzyme.Microb.Technol., 130, 2019
|
|
2ZJD
 
 | | Crystal Structure of LC3-p62 complex | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B precursor, undecameric peptide from Sequestosome-1 | | Authors: | Ichimura, Y, Kumanomidou, T, Sou, Y, Mizushima, T, Ezaki, J, Ueno, T, Kominami, E, Yamane, T, Tanaka, K, Komatsu, M. | | Deposit date: | 2008-03-05 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Basis for Sorting Mechanism of p62 in Selective Autophagy
J.Biol.Chem., 283, 2008
|
|
2ZQA
 
 | | Cefotaxime acyl-intermediate structure of class a beta-lacta Toho-1 E166A/R274N/R276N triple mutant | | Descriptor: | Beta-lactamase Toho-1, CEFOTAXIME, C3' cleaved, ... | | Authors: | Nitanai, Y, Shimamura, T, Uchiyama, T, Ishii, Y, Takehira, M, Yutani, K, Matsuzawa, H, Miyano, M. | | Deposit date: | 2008-08-07 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural Basis of Extend Spectrum Beta-Lactamase in Correlation of Enzymatic Kinetics and Thermal Stability of Acyl-Intermediates
To be Published
|
|
2ZQD
 
 | | Ceftazidime acyl-intermediate structure of class a beta-lact Toho-1 E166A/R274N/R276N triple mutant | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase Toho-1, SULFATE ION | | Authors: | Nitanai, Y, Shimamura, T, Uchiyama, T, Ishii, Y, Takehira, M, Yutani, K, Matsuzawa, H, Miyano, M. | | Deposit date: | 2008-08-07 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural Basis of Extend Spectrum Beta-Lactamase in Correlation of Enzymatic Kinetics and Thermal Stability of Acyl-Intermediates
To be Published
|
|
2ZOM
 
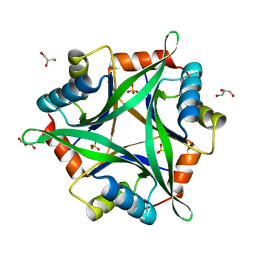 | | Crystal structure of CutA1 from Oryza sativa | | Descriptor: | GLYCEROL, Protein CutA, chloroplast, ... | | Authors: | Kezuka, Y, Bagautdinov, B, Katoh, S, Ohtake, Y, Yutani, K, Nonaka, T, Katoh, E. | | Deposit date: | 2008-05-23 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Crystal structure of CutA1 from Oryza sativa
To be Published
|
|
3ASE
 
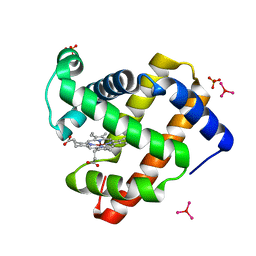 | | Crystal Structure of Zinc myoglobin soaked with Ru3O cluster | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING ZN, SULFATE ION, ... | | Authors: | Koshiyama, T, Shirai, M, Hikage, T, Tabe, H, Tanaka, K, Kitagawa, S, Ueno, T. | | Deposit date: | 2010-12-11 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Post-Crystal Engineering of Zinc-Substituted Myoglobin to Construct a Long-Lived Photoinduced Charge-Separation System
Angew.Chem.Int.Ed.Engl., 2011
|
|
2ZQC
 
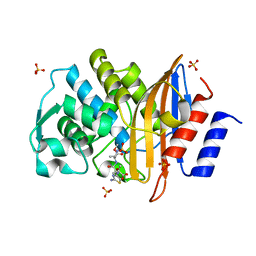 | | Aztreonam acyl-intermediate structure of class a beta-lactam Toho-1 E166A/R274N/R276N triple mutant | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, Beta-lactamase Toho-1, SULFATE ION | | Authors: | Nitanai, Y, Shimamura, T, Uchiyama, T, Ishii, Y, Takehira, M, Yutani, K, Matsuzawa, H, Miyano, M. | | Deposit date: | 2008-08-07 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Structural Basis of Extend Spectrum Beta-Lactamase in Correlation of Enzymatic Kinetics and Thermal Stability of Acyl-Intermediates
To be Published
|
|
2YVC
 
 | |
2Z9W
 
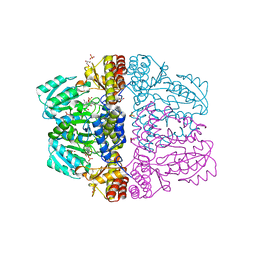 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxal | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, Aspartate aminotransferase, GLYCEROL, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
6L9C
 
 | | Neutron structure of copper amine oxidase from Arthrobacter glibiformis at pD 7.4 | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Kurihara, K, Shoji, M, Shibazaki, C, Sunami, T, Tamada, T, Yano, N, Yamada, T, Kusaka, K, Suzuki, M, Shigeta, Y, Kuroki, R, Hayashi, H, Yano, Y, Tanizawa, K, Adachi, M, Okajima, T. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.14 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography of copper amine oxidase reveals keto/enolate interconversion of the quinone cofactor and unusual proton sharing.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2XZB
 
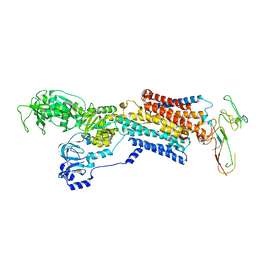 | | Pig Gastric H,K-ATPase with bound BeF and SCH28080 | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Fujiyoshi, Y. | | Deposit date: | 2010-11-24 | | Release date: | 2011-01-26 | | Last modified: | 2020-09-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (7 Å) | | Cite: | Conformational Rearrangement of Gastric H(+),K(+)- ATPase Induced by an Acid Suppressant.
Nat.Commun., 2, 2011
|
|
2ZPH
 
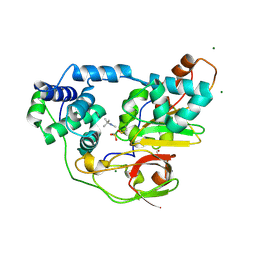 | | Complex of Fe-type nitrile hydratase with tert-butylisonitrile, photo-activated for 340min at 293K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
2Z9U
 
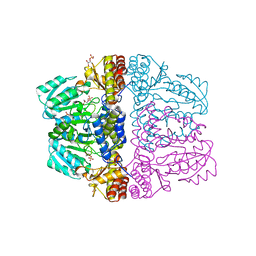 | | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti at 2.0 A resolution | | Descriptor: | Aspartate aminotransferase, GLYCEROL, SULFATE ION | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
2ZPB
 
 | | nitrosylated Fe-type nitrile hydratase | | Descriptor: | FE (III) ION, MAGNESIUM ION, NITRIC OXIDE, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-09 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
2ZPI
 
 | | Complex of Fe-type nitrile hydratase with tert-butylisonitrile, photo-activated for 440min at 293K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
2YN9
 
 | | Cryo-EM structure of gastric H+,K+-ATPase with bound rubidium | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Friedrich, T, Fujiyoshi, Y. | | Deposit date: | 2012-10-13 | | Release date: | 2012-11-07 | | Last modified: | 2014-07-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (8 Å) | | Cite: | Cryo-Em Structure of Gastric H+,K+-ATPase with a Single Occupied Cation-Binding Site.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
