1XKM
 
 | | NMR structure of antimicrobial peptide distinctin in water | | Descriptor: | Distinctin chain A, Distinctin chain B | | Authors: | Amodeo, P, Raimondo, D, Andreotti, G, Motta, A, Scaloni, A. | | Deposit date: | 2004-09-29 | | Release date: | 2005-04-05 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | A folding-dependent mechanism of antimicrobial peptide resistance to degradation unveiled by solution structure of distinctin.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
9CMC
 
 | | Crystal structure of the peanut allergen Ara h 2 with two human derived Fab antibodies 22S1 and 23P34 | | Descriptor: | Ara h 2 allergen, Fab 22S1 heavy chain, Fab 22S1 light chain, ... | | Authors: | Pedersen, L.C, Mueller, G.A, Min, J. | | Deposit date: | 2024-07-14 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Germline-encoded recognition of peanut underlies development of convergent antibodies in humans.
Sci Transl Med, 17, 2025
|
|
6LLW
 
 | |
6X5T
 
 | | Human Alpha-1,6-fucosyltransferase (FUT8) bound to GDP and A3-Asn | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-(1,6)-fucosyltransferase, GLYCEROL, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Characterizing human alpha-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases.
J.Biol.Chem., 295, 2020
|
|
7NYJ
 
 | | Structure of OBP1 from Varroa destructor, form P3<2>21 | | Descriptor: | CALCIUM ION, Odorant Binding Protein 1 from Varoa destructor, form P3<2>21, ... | | Authors: | Cambillau, C, Amigues, B, Roussel, A, Leone, P, Gaubert, A, Pelosi, P. | | Deposit date: | 2021-03-22 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A new non-classical fold of varroa odorant-binding proteins reveals a wide open internal cavity.
Sci Rep, 11, 2021
|
|
7NZA
 
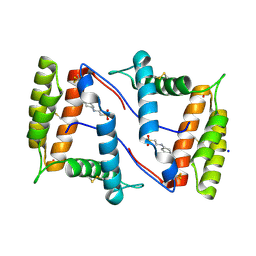 | | Structure of OBP1 from Varroa destructor, form P2<1> | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Odorant Binding Protein from Varroa destructor, form P2<1>, ... | | Authors: | Cambillau, C, Amigues, B, Roussel, A, Leone, P, Gaubert, A, Pelosi, P. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | A new non-classical fold of varroa odorant-binding proteins reveals a wide open internal cavity.
Sci Rep, 11, 2021
|
|
8G9E
 
 | |
6X5R
 
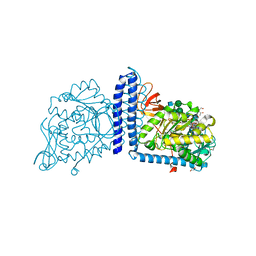 | | Human Alpha-1,6-fucosyltransferase (FUT8) bound to GDP and A2-Asn | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A2-Asn, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterizing human alpha-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases.
J.Biol.Chem., 295, 2020
|
|
6X5H
 
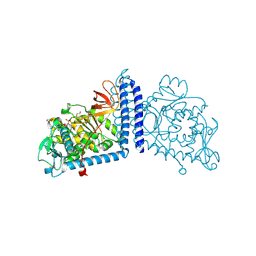 | | Human Alpha-1,6-fucosyltransferase (FUT8) bound to GDP | | Descriptor: | 1,2-ETHANEDIOL, Alpha-(1,6)-fucosyltransferase, GLYCEROL, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterizing human alpha-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases.
J.Biol.Chem., 295, 2020
|
|
6X5U
 
 | | Human Alpha-1,6-fucosyltransferase (FUT8) bound to GDP and NM5M2-Asn | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Characterizing human alpha-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases.
J.Biol.Chem., 295, 2020
|
|
6X5S
 
 | | Human Alpha-1,6-fucosyltransferase (FUT8) bound to GDP and A3'-Asn | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A3'-Asn, Alpha-(1,6)-fucosyltransferase, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Characterizing human alpha-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases.
J.Biol.Chem., 295, 2020
|
|
7MOF
 
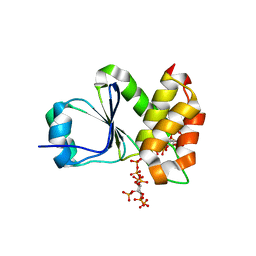 | | Crystal Structure of Arabidopsis thaliana Plant and Fungi Atypical Dual Specificity Phosphatase 1(AtPFA-DSP1 ) Cys150Ser in complex with 6-diphosphoinositol 1,2,3,4,5-pentakisphosphate 6-InsP7 | | Descriptor: | (1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, INOSITOL HEXAKISPHOSPHATE, Tyrosine-protein phosphatase DSP1 | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2021-05-01 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A structural expose of noncanonical molecular reactivity within the protein tyrosine phosphatase WPD loop.
Nat Commun, 13, 2022
|
|
7MOJ
 
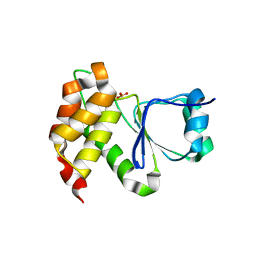 | |
7MOL
 
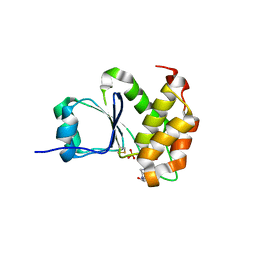 | |
7MOM
 
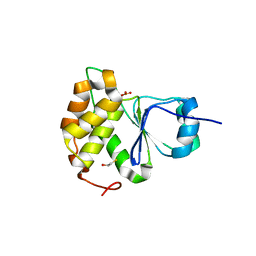 | |
7MOG
 
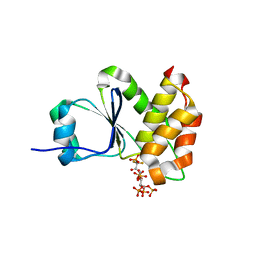 | | Crystal Structure of Arabidopsis thaliana Plant and Fungi Atypical Dual Specificity Phosphatase 1(AtPFA-DSP1 ) Cys150Ser in complex with 5-PCF2 Am-InsP5, an analogue of 5-InsP7 | | Descriptor: | (1,1-difluoro-2-oxo-2-{[(1s,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl]amino}ethyl)phosphonic acid, Tyrosine-protein phosphatase DSP1 | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2021-05-01 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural expose of noncanonical molecular reactivity within the protein tyrosine phosphatase WPD loop.
Nat Commun, 13, 2022
|
|
7MOH
 
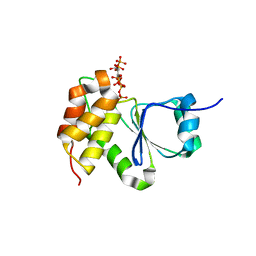 | | Crystal Structure of Arabidopsis thaliana Plant and Fungi Atypical Dual Specificity Phosphatase 1(AtPFA-DSP1 ) Cys150Ser in complex with 5-diphosphoinositol 1,3,4,6-tetrakisphosphate (5PP-InsP4) and phosphate in conformation B (Pi(B)) | | Descriptor: | (1r,2R,3S,4r,5R,6S)-4-hydroxy-2,3,5,6-tetrakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, PHOSPHATE ION, Tyrosine-protein phosphatase DSP1 | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2021-05-01 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural expose of noncanonical molecular reactivity within the protein tyrosine phosphatase WPD loop.
Nat Commun, 13, 2022
|
|
7MOD
 
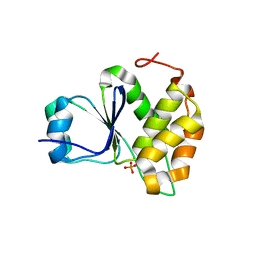 | |
7MOE
 
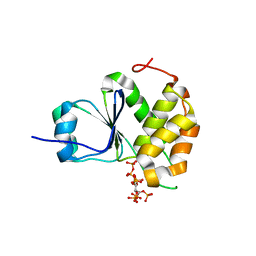 | | Crystal Structure of Arabidopsis thaliana Plant and Fungi Atypical Dual Specificity Phosphatase 1(AtPFA-DSP1)Cys150Ser in complex with 5-diphosphoinositol 1,2,3,4,6-pentakisphosphate (5-InsP7) | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, Tyrosine-protein phosphatase DSP1 | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2021-05-01 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structural expose of noncanonical molecular reactivity within the protein tyrosine phosphatase WPD loop.
Nat Commun, 13, 2022
|
|
7MOK
 
 | |
7MOI
 
 | |
9EXA
 
 | |
