1RCS
 
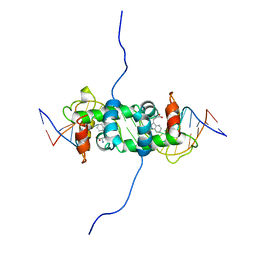 | | NMR STUDY OF TRP REPRESSOR-OPERATOR DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*TP*AP*GP*TP*AP*CP*G)-3'), TRP REPRESSOR, TRYPTOPHAN | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the trp repressor-operator DNA complex.
J.Mol.Biol., 238, 1994
|
|
1WRT
 
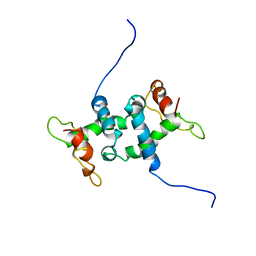 | | NMR STUDY OF APO TRP REPRESSOR | | Descriptor: | APO TRP REPRESSOR | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structures of the Escherichia coli trp holo- and aporepressor.
J.Mol.Biol., 229, 1993
|
|
1WRS
 
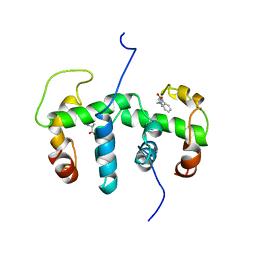 | | NMR STUDY OF HOLO TRP REPRESSOR | | Descriptor: | HOLO TRP REPRESSOR, TRYPTOPHAN | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structures of the Escherichia coli trp holo- and aporepressor.
J.Mol.Biol., 229, 1993
|
|
7X7T
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zheng, Q, Li, S, Zhang, T, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X7U
 
 | | Cryo-EM structure of SARS-CoV-2 Delta variant spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X7V
 
 | | Cryo-EM structure of SARS-CoV spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XDS
 
 | |
7XE0
 
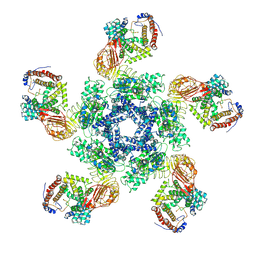 | | Cryo-EM structure of plant NLR Sr35 resistosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AvrSr35, Sr35 | | Authors: | Ouyang, S.Y, Zhao, Y.B, Li, Z.K, Liu, M.X. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Pathogen effector AvrSr35 triggers Sr35 resistosome assembly via a direct recognition mechanism.
Sci Adv, 8, 2022
|
|
8Y83
 
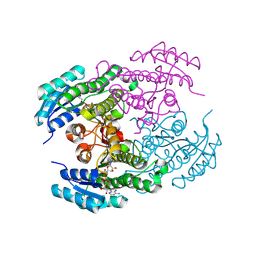 | | Crystal structure of a ketoreductase from Sphingobacterium siyangense SY1 with co-enzyme | | Descriptor: | NAD(P)-dependent dehydrogenase (Short-subunit alcohol dehydrogenase family), NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zheng, Z.R, Wei, H.L, Liu, W.D, You, S. | | Deposit date: | 2024-02-05 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based reshaping of a new ketoreductase from Sphingobacterium siyangense SY1 toward alpha-haloacetophenones.
Int.J.Biol.Macromol., 277, 2024
|
|
8Y7R
 
 | |
7XVG
 
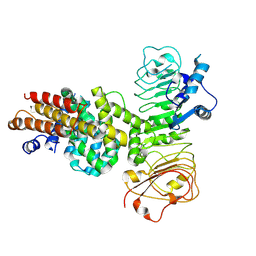 | | Cryo-EM structure of binary complex of plant NLR Sr35 and effector AvrSr35 | | Descriptor: | AvrSr35, Sr35 | | Authors: | Ouyang, S.Y, Zhao, Y.B, Li, Z.K, Liu, M.X. | | Deposit date: | 2022-05-23 | | Release date: | 2022-09-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Pathogen effector AvrSr35 triggers Sr35 resistosome assembly via a direct recognition mechanism.
Sci Adv, 8, 2022
|
|
7XX2
 
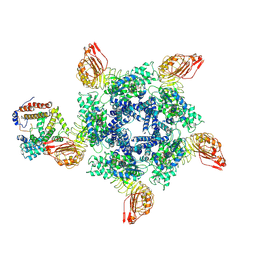 | | Cryo-EM structure of Sr35 resistosome induced by AvrSr35 R381A | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AvrSr35, CNL9 | | Authors: | Ouyang, S.Y, Zhao, Y.B, Li, Z.K, Liu, M.X. | | Deposit date: | 2022-05-28 | | Release date: | 2022-11-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Pathogen effector AvrSr35 triggers Sr35 resistosome assembly via a direct recognition mechanism.
Sci Adv, 8, 2022
|
|
8HTR
 
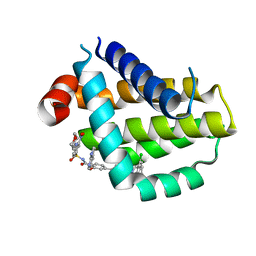 | | Crystal structure of Bcl2 in complex with S-9c | | Descriptor: | 4-[4-[(2~{S})-2-(2-chlorophenyl)pyrrolidin-1-yl]phenyl]-~{N}-[3-nitro-4-(oxan-4-ylmethylamino)phenyl]sulfonyl-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide, Apoptosis regulator Bcl-2 | | Authors: | Liu, J, Xu, M, Feng, Y, Liu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of the Clinical Candidate Sonrotoclax (BGB-11417), a Highly Potent and Selective Inhibitor for Both WT and G101V Mutant Bcl-2.
J.Med.Chem., 67, 2024
|
|
8HTS
 
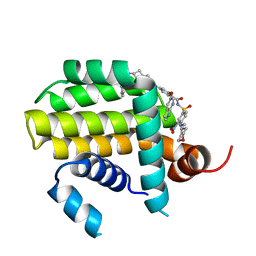 | | Crystal structure of Bcl2 in complex with S-10r | | Descriptor: | 4-[2-[(2~{S})-2-(2-cyclopropylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-~{N}-[3-nitro-4-(oxan-4-ylmethylamino)phenyl]sulfonyl-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide, Apoptosis regulator Bcl-2 | | Authors: | Liu, J, Xu, M, Feng, Y, Liu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of the Clinical Candidate Sonrotoclax (BGB-11417), a Highly Potent and Selective Inhibitor for Both WT and G101V Mutant Bcl-2.
J.Med.Chem., 67, 2024
|
|
7D6N
 
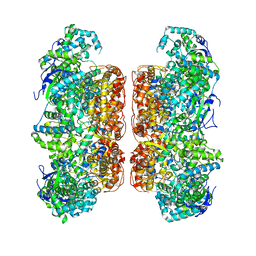 | |
7D6M
 
 | |
7WZW
 
 | | Cryo-EM structure of MEC1-DDC2-MMS | | Descriptor: | DNA damage checkpoint protein LCD1, Serine/threonine-protein kinase MEC1 | | Authors: | Zhang, Q, Zhang, Q. | | Deposit date: | 2022-02-19 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of Mec1/ATR kinase endogenously stimulated by different genotoxins.
Cell Discov, 8, 2022
|
|
7WZR
 
 | | Cryo-EM structure of Mec1-HU | | Descriptor: | DNA damage checkpoint protein LCD1, Serine/threonine-protein kinase MEC1 | | Authors: | Zhang, Q, Zhang, Q. | | Deposit date: | 2022-02-19 | | Release date: | 2023-03-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structures of Mec1/ATR kinase endogenously stimulated by different genotoxins.
Cell Discov, 8, 2022
|
|
8Y9O
 
 | | Crystal structure of AtKAI2 in complex with KK181N1 | | Descriptor: | (7-methyl-2,3-dihydroindol-1-yl)-(1,2,3-triazol-1-yl)methanone, Probable esterase KAI2 | | Authors: | Wang, J.W, Miyakawa, T, Asami, T. | | Deposit date: | 2024-02-07 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and structure-guided development of triazole urea-based selective antagonists of Arabidopsis karrikin signaling.
Nat Commun, 16, 2025
|
|
6IWF
 
 | | Crystal structure of HitA from Pseudomonas aeruginosa | | Descriptor: | Ferric iron-binding protein HitA | | Authors: | Guo, Y, Zhengrui, Z, Li, H. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.70662332 Å) | | Cite: | Identification and Characterization of a Metalloprotein Involved in Gallium Internalization in Pseudomonas aeruginosa.
Acs Infect Dis., 5, 2019
|
|
