1XMA
 
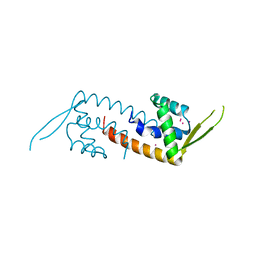 | | Structure of a transcriptional regulator from Clostridium thermocellum Cth-833 | | Descriptor: | MERCURY (II) ION, Predicted transcriptional regulator, UNKNOWN ATOM OR ION | | Authors: | Yang, H, Chen, L, Lee, D, Habel, J, Nguyen, J, Chang, S.-H, Kataeva, I, Xu, H, Chang, J, Zhao, M, Horanyi, P, Florence, Q, Zhou, W, Tempel, W, Lin, D, Praissman, J, Zhang, H, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Ljungdahl, L, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-10-01 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure of a transcriptional regulator from Clostridium thermocellum Cth-833
To be published
|
|
6K7C
 
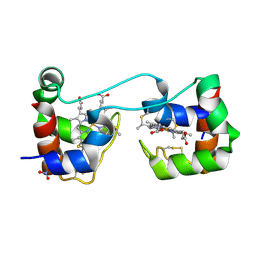 | | Dimeric Shewanella violacea cytochrome c5 | | Descriptor: | HEME C, NITRATE ION, Soluble cytochrome cA | | Authors: | Yang, H, Yamanaka, M, Nagao, S, Yasuhara, K, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2019-06-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Protein surface charge effect on 3D domain swapping in cells for c-type cytochromes.
Biochim Biophys Acta Proteins Proteom, 1867, 2019
|
|
1WOF
 
 | | Crystal Structure Of SARS-CoV Mpro in Complex with an Inhibitor N1 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]-L-ALANYL-L-VALYL-N~1~-((1S)-4-ETHOXY-4-OXO-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Yang, H, Bartlam, M, Xue, X, Yang, K, Liang, W, Rao, Z. | | Deposit date: | 2004-08-18 | | Release date: | 2005-08-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases.
Plos Biol., 3, 2005
|
|
7DL2
 
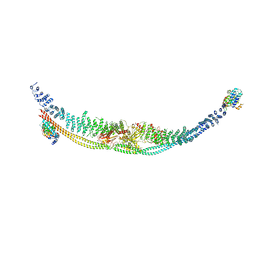 | | Cryo-EM structure of human TSC complex | | Descriptor: | Hamartin, Isoform 7 of Tuberin, TBC1 domain family member 7, ... | | Authors: | Yang, H, Yu, Z, Chen, X, Li, J, Li, N, Cheng, J, Gao, N, Yuan, H, Ye, D, Guan, K, Xu, Y. | | Deposit date: | 2020-11-25 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into TSC complex assembly and GAP activity on Rheb.
Nat Commun, 12, 2021
|
|
7UU4
 
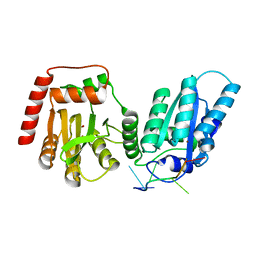 | | Crystal structure of APOBEC3G complex with ssRNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, RNA (5'-R(P*UP*AP*AP*UP*UP*U)-3'), SULFATE ION, ... | | Authors: | Yang, H, Li, S, Chen, X.S. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G.
Nat Commun, 13, 2022
|
|
7UU5
 
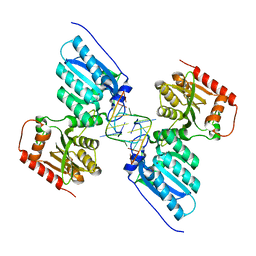 | | Crystal structure of APOBEC3G complex with 5'-Overhang dsRNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, RNA (5'-R(P*UP*AP*AP*CP*CP*GP*CP*AP*GP*CP*G)-3'), RNA (5'-R(P*UP*AP*AP*CP*GP*CP*UP*GP*CP*GP*G)-3'), ... | | Authors: | Yang, H, Li, S, Chen, X.S. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G.
Nat Commun, 13, 2022
|
|
7UU3
 
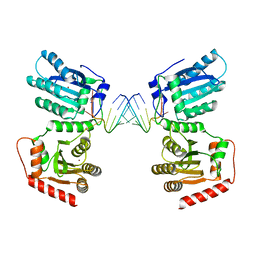 | | Crystal structure of APOBEC3G complex with 3'overhangs RNA-Complex | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, RNA (5'-R(*CP*CP*CP*AP*CP*GP*GP*GP*AP*AP*U)-3'), RNA (5'-R(*CP*CP*CP*GP*UP*GP*GP*GP*AP*AP*U)-3'), ... | | Authors: | Yang, H, Li, S, Chen, X.S. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G.
Nat Commun, 13, 2022
|
|
7D2T
 
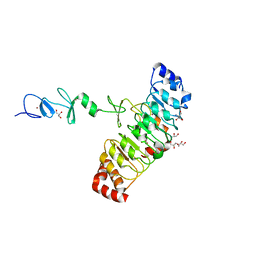 | | Crystal structure of Rsu1/PINCH1_LIM45C complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, ... | | Authors: | Yang, H, Wei, Z, Yu, C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
7D2S
 
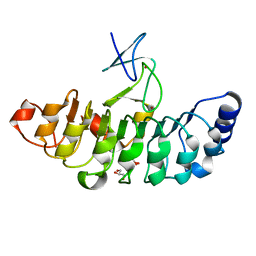 | | Crystal structure of Rsu1/PINCH1_LIM5C complex | | Descriptor: | GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, Ras suppressor protein 1, ... | | Authors: | Yang, H, Wei, Z, Cong, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
7D2U
 
 | | Crystal structure of Rsu1/PINCH1_LIM45C complex | | Descriptor: | GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, MALONATE ION, ... | | Authors: | Yang, H, Wei, Z, Cong, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
1YEM
 
 | | Conserved hypothetical protein Pfu-838710-001 from Pyrococcus furiosus | | Descriptor: | Conserved hypothetical protein Pfu-838710-001, PLATINUM (II) ION, UNKNOWN ATOM OR ION | | Authors: | Yang, H, Chang, J, Shah, A, Ng, J.D, Liu, Z.-J, Chen, L, Lee, D, Tempel, W, Praissman, J.L, Lin, D, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-28 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved hypothetical protein Pfu-838710-001 from Pyrococcus furiosus
To be published
|
|
6D7C
 
 | |
6D7U
 
 | |
6D8B
 
 | |
6D8D
 
 | |
5U33
 
 | | Crystal structure of AacC2c1-sgRNA-extended non-target DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease C2c1, Non-target DNA strand, SULFATE ION, ... | | Authors: | Yang, H, Gao, P, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease.
Cell, 167, 2016
|
|
5VW1
 
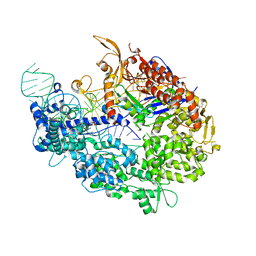 | | Crystal structure of SpyCas9-sgRNA-AcrIIA4 ternary complex | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, H, Patel, D.J. | | Deposit date: | 2017-05-21 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Inhibition Mechanism of an Anti-CRISPR Suppressor AcrIIA4 Targeting SpyCas9.
Mol. Cell, 67, 2017
|
|
4W8N
 
 | | The crystal structure of hemagglutinin from a swine influenza virus (A/swine/Missouri/2124514/2006) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yang, H, Carney, P.J, Tumpey, T.M, Stevens, J. | | Deposit date: | 2014-08-25 | | Release date: | 2015-02-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Assessment of transmission, pathogenesis and adaptation of H2 subtype influenza viruses in ferrets.
Virology, 477C, 2015
|
|
6N4D
 
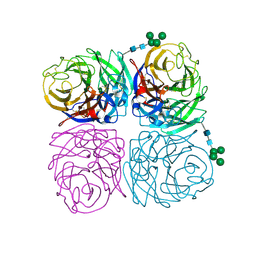 | | The crystal structure of neuramindase from A/canine/IL/11613/2015 (H3N2) influenza virus. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yang, H, Stevens, J. | | Deposit date: | 2018-11-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Assessment of Molecular, Antigenic, and Pathological Features of Canine Influenza A(H3N2) Viruses That Emerged in the United States.
J. Infect. Dis., 216, 2017
|
|
6N4F
 
 | |
6N6B
 
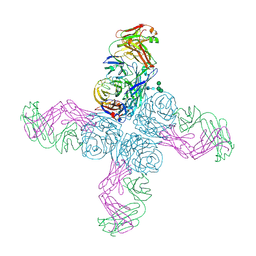 | |
2H2Z
 
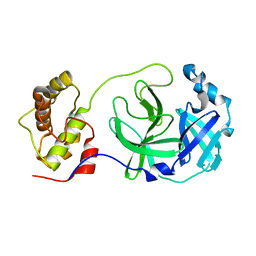 | | Crystal structure of SARS-CoV main protease with authentic N and C-termini | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Yang, H, Xue, X, Shen, W, Zhao, Q, Rao, Z. | | Deposit date: | 2006-05-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction
J.Mol.Biol., 366, 2007
|
|
8HWN
 
 | | aldo-keto reductase DepB | | Descriptor: | CHLORIDE ION, DepB, SODIUM ION | | Authors: | Chen, M, Yang, H, Lu, F. | | Deposit date: | 2022-12-31 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of DepB capable of DON detoxification
To Be Published
|
|
4JT6
 
 | | structure of mTORDeltaN-mLST8-PI-103 complex | | Descriptor: | 3-(4-MORPHOLIN-4-YLPYRIDO[3',2':4,5]FURO[3,2-D]PYRIMIDIN-2-YL)PHENOL, mLST8, mTOR | | Authors: | Pavletich, N.P, Yang, H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | mTOR kinase structure, mechanism and regulation.
Nature, 497, 2013
|
|
4JSX
 
 | | structure of mTORDeltaN-mLST8-Torin2 complex | | Descriptor: | 9-(6-aminopyridin-3-yl)-1-[3-(trifluoromethyl)phenyl]benzo[h][1,6]naphthyridin-2(1H)-one, Serine/threonine-protein kinase mTOR, Target of rapamycin complex subunit LST8 | | Authors: | Pavletich, N.P, Yang, H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | mTOR kinase structure, mechanism and regulation.
Nature, 497, 2013
|
|
