8WED
 
 | | Crystal structure of Arabidopsis thaliana MIK2 ectodomain in complex with BAK1 ectodomain and Fusarium oxysporum SCOOPL | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wan, L.H, Hu, Y.X, Wu, H.M. | | Deposit date: | 2023-09-17 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation.
Nat.Plants, 10, 2024
|
|
4BA6
 
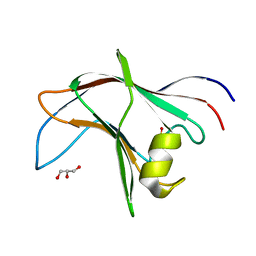 | | High Resolution structure of the C-terminal family 65 Carbohydrate Binding Module (CBM65B) of endoglucanase Cel5A from Eubacterium cellulosolvens | | Descriptor: | Endoglucanase cel5A, GLYCEROL | | Authors: | Venditto, I, Luis, A.S, Basle, A, Temple, M, Ferreira, L.M.A, Fontes, C.M.G.A, Gilbert, H.J, Najmudin, S. | | Deposit date: | 2012-09-11 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Understanding how noncatalytic carbohydrate binding modules can display specificity for xyloglucan.
J. Biol. Chem., 288, 2013
|
|
6AC0
 
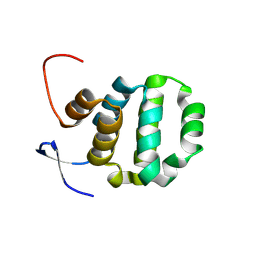 | | Crystal structure of TRADD death domain GlcNAcylated by EPEC effector NleB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor receptor type 1-associated DEATH domain protein | | Authors: | Ding, J, Shao, F. | | Deposit date: | 2018-07-24 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structural and Functional Insights into Host Death Domains Inactivation by the Bacterial Arginine GlcNAcyltransferase Effector.
Mol.Cell, 74, 2019
|
|
6AC5
 
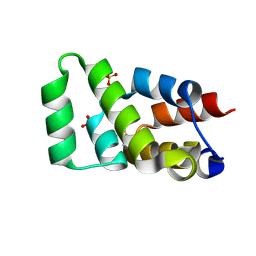 | | Crystal structure of RIPK1 death domain GlcNAcylated by EPEC effector NleB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-interacting serine/threonine-protein kinase 1, SULFATE ION | | Authors: | Ding, J, Shao, F. | | Deposit date: | 2018-07-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural and Functional Insights into Host Death Domains Inactivation by the Bacterial Arginine GlcNAcyltransferase Effector.
Mol.Cell, 74, 2019
|
|
6ACI
 
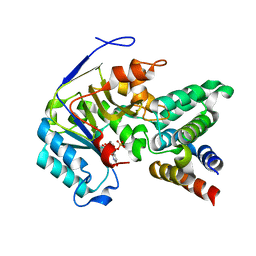 | | Crystal structure of EPEC effector NleB in complex with FADD death domain | | Descriptor: | FAS-associated death domain protein, MANGANESE (II) ION, T3SS secreted effector NleB homolog, ... | | Authors: | Ding, J, Shao, F. | | Deposit date: | 2018-07-26 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Functional Insights into Host Death Domains Inactivation by the Bacterial Arginine GlcNAcyltransferase Effector.
Mol.Cell, 74, 2019
|
|
8WCS
 
 | | Cryo-EM structure of Cas13h1-crRNA binary complex | | Descriptor: | 66-nt crRNA, Cas13h1, MAGNESIUM ION | | Authors: | Zhang, C. | | Deposit date: | 2023-09-13 | | Release date: | 2024-05-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism for target RNA recognition and cleavage of Cas13h.
Nucleic Acids Res., 52, 2024
|
|
8WCE
 
 | | Cryo-EM structure of a protein-RNA complex | | Descriptor: | MAGNESIUM ION, RNA (31-MER), RNA (66-MER), ... | | Authors: | Li, Z. | | Deposit date: | 2023-09-11 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Molecular mechanism for target RNA recognition and cleavage of Cas13h.
Nucleic Acids Res., 52, 2024
|
|
7RA0
 
 | | LC3A in complex with Fragment 2-10 | | Descriptor: | (5-ethyl-2-methyl-1H-indol-3-yl)acetic acid, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Rouge, L, Steffek, M, Helgason, E, Dueber, E, Mulvihill, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A Multifaceted Hit-Finding Approach Reveals Novel LC3 Family Ligands.
Biochemistry, 62, 2023
|
|
7R9W
 
 | | LC3A in complex with Fragment 1-1 | | Descriptor: | 4-phenoxybenzoic acid, GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Rouge, L, Steffek, M, Helgason, E, Dueber, E, Mulvihill, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Multifaceted Hit-Finding Approach Reveals Novel LC3 Family Ligands.
Biochemistry, 62, 2023
|
|
7R9Z
 
 | | LC3A in complex with Fragment 2-3 | | Descriptor: | (5-fluoro-1H-indol-3-yl)acetic acid, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Rouge, L, Steffek, M, Helgason, E, Dueber, E, Mulvihill, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A Multifaceted Hit-Finding Approach Reveals Novel LC3 Family Ligands.
Biochemistry, 62, 2023
|
|
8WEB
 
 | | Crystal structure of Arabidopsis thaliana MIK2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MDIS1-interacting receptor like kinase 2 | | Authors: | Wan, L.H, Hu, Y.X, Wu, H.M. | | Deposit date: | 2023-09-17 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation.
Nat.Plants, 10, 2024
|
|
8WEF
 
 | | Crystal structure of Brassica napus MIK2 ectodomain in complex with Fusarium oxysporum SCOOPL | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wan, L.H, Hu, Y.X, Wu, H.M. | | Deposit date: | 2023-09-17 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation.
Nat.Plants, 10, 2024
|
|
8WEI
 
 | | Crystal structure of Brassica napus MIK2 ectodomain (N393D mutant) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wan, L.H, Hu, Y.X, Wu, H.M. | | Deposit date: | 2023-09-17 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation.
Nat.Plants, 10, 2024
|
|
8WEG
 
 | | Crystal structure of Brassica napus MIK2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wan, L.H, Hu, Y.X, Wu, H.M. | | Deposit date: | 2023-09-17 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation.
Nat.Plants, 10, 2024
|
|
8WEH
 
 | | Crystal structure of Brassica napus MIK2 ectodomain (N393A mutant) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wan, L.H, Hu, Y.X, Wu, H.M. | | Deposit date: | 2023-09-17 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation.
Nat.Plants, 10, 2024
|
|
8WEE
 
 | | Crystal structure of Arabidopsis thaliana MIK2 ectodomain in complex with SCOOP12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wan, L.H, Hu, Y.X, Wu, H.M. | | Deposit date: | 2023-09-17 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation.
Nat.Plants, 10, 2024
|
|
6B4U
 
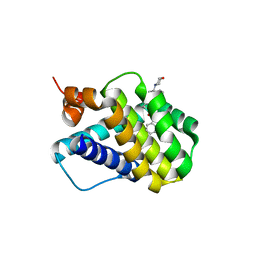 | | Crystal structure of MCL-1 in complex with a BIM competitive inhibitor | | Descriptor: | 7-(2-methylphenyl)-1-[2-(morpholin-4-yl)ethyl]-3-{3-[(naphthalen-1-yl)oxy]propyl}-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Judge, R.A, Souers, A.J. | | Deposit date: | 2017-09-27 | | Release date: | 2017-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity.
J. Med. Chem., 58, 2015
|
|
6B4L
 
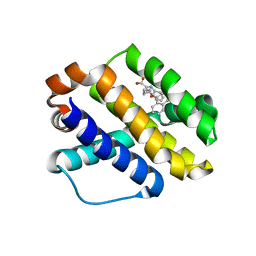 | |
7END
 
 | |
7EN9
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-02 | | Descriptor: | 3C-like proteinase, 5-bromanyl-~{N}-methyl-3-nitro-2-[(4~{R},5~{S})-2-(7-oxidanylisoquinolin-4-yl)carbonyl-4-phenyl-2,7-diazaspiro[4.4]nonan-7-yl]benzamide | | Authors: | Hou, N, Peng, C, Hu, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
7EN8
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-04 | | Descriptor: | 3C-like proteinase, GLYCEROL, ~{N}-[(1~{S},2~{R})-2-[[4-bromanyl-2-(methylcarbamoyl)-6-nitro-phenyl]amino]cyclohexyl]isoquinoline-4-carboxamide | | Authors: | Hou, N, Peng, C, Hu, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
7ENE
 
 | |
6KTW
 
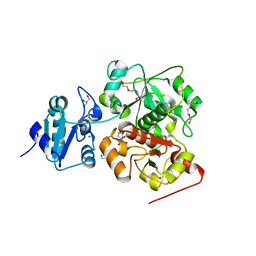 | | structure of EanB with hercynine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-29 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.931 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
6KU2
 
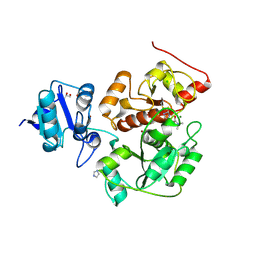 | | The structure of EanB/Y353A complex with ergothioneine covalent linked with persulfide Cys412 | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
6KTZ
 
 | | The complex structure of EanB/C412S with hercynine | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-29 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
