4E1R
 
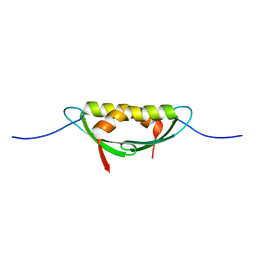 | |
4E1P
 
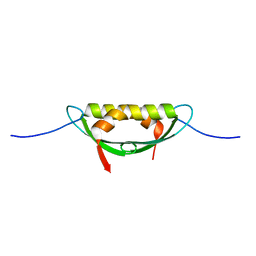 | |
4WG0
 
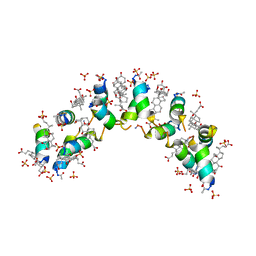 | |
1UNM
 
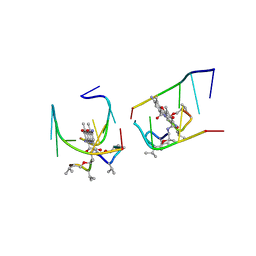 | | Crystal structure of 7-Aminoactinomycin D with non-complementary DNA | | Descriptor: | 5'-D(*TP*TP*AP*GP*BRU*TP)-3', 7-AMINOACTINOMYCIN D | | Authors: | Alexopoulos, E.C, Klement, R, Jares-Erijman, E.A, Uson, I, Jovin, T.M, Sheldrick, G.M. | | Deposit date: | 2003-09-11 | | Release date: | 2004-09-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal and Solution Structures of 7-Amino-Actinomycin D Complexes with D(Ttagbrut), D(Ttagtt) and D(Tttagttt)
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5LCY
 
 | | Formaldehyde-Responsive Regulator FrmR E64H variant from Salmonella enterica serovar Typhimurium | | Descriptor: | Frmr | | Authors: | Pohl, E, Robinson, N, Osman, D, Piergentili, C, Uson, I. | | Deposit date: | 2016-06-22 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Effectors and Sensory Sites of Formaldehyde-responsive Regulator FrmR and Metal-sensing Variant.
J.Biol.Chem., 291, 2016
|
|
5M2Z
 
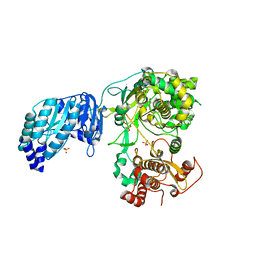 | | Crystal structure of the full-length Zika virus NS5 protein (Human isolate Z1106033) | | Descriptor: | NS5, PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Ferrero, D.S, Ruiz-Arroyo, V.M, Soler, N, Uson, I, Verdaguer, N. | | Deposit date: | 2016-10-13 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Supramolecular arrangement of the full-length Zika virus NS5.
Plos Pathog., 15, 2019
|
|
1W7Z
 
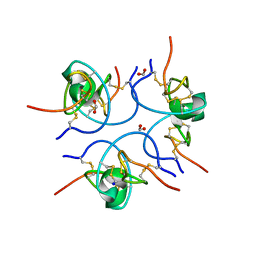 | | Crystal structure of the free (uncomplexed) Ecballium elaterium trypsin inhibitor (EETI-II) | | Descriptor: | FORMIC ACID, SODIUM ION, TRYPSIN INHIBITOR II | | Authors: | Kraetzner, R, Debreczeni, J.E, Pape, T, Kolmar, H, Schneider, T.R, Uson, I, Scheldrick, G.M. | | Deposit date: | 2004-09-14 | | Release date: | 2005-11-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of Ecballium Elaterium Trypsin Inhibitor II (Eeti-II): A Rigid Molecular Scaffold
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5M2X
 
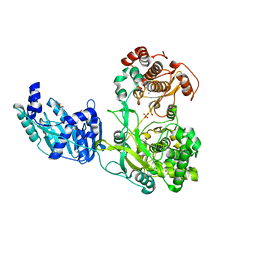 | | Crystal structure of the full-length Zika virus NS5 protein (Human isolate Z1106033) | | Descriptor: | NS5, PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Ferrero, D.S, Ruiz-Arroyo, V.M, Soler, N, Uson, I, Verdaguer, N. | | Deposit date: | 2016-10-13 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.991 Å) | | Cite: | Supramolecular arrangement of the full-length Zika virus NS5.
Plos Pathog., 15, 2019
|
|
1UNJ
 
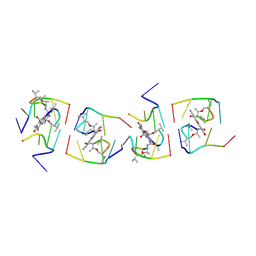 | | Crystal structure of a 7-Aminoactinomycin D complex with non-complementary DNA | | Descriptor: | 5'-D(*TP*TP*AP*GP*BRU*TP)-3', 7-AMINO-ACTINOMYCIN D | | Authors: | Alexopoulos, E.C, Klement, R, Jares-Erijman, E.A, Uson, I, Jovin, T.M, Sheldrick, G.M. | | Deposit date: | 2003-09-10 | | Release date: | 2004-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal and Solution Structures of 7-Amino-Actinomycin D Complexes with D(Ttagbrut), D(Ttagtt) and D(Tttagttt)
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5MUN
 
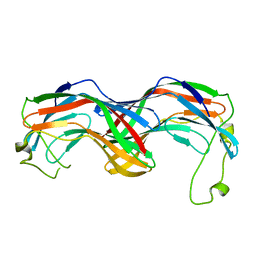 | | Structural insight into zymogenic latency of gingipain K from Porphyromonas gingivalis. | | Descriptor: | AZIDE ION, Lys-gingipain W83 | | Authors: | Pomowski, A, Uson, I, Nowakovska, Z, Veillard, F, Sztukowska, M.N, Guevara, T, Goulas, T, Mizgalska, D, Nowak, M, Potempa, B, Huntington, J.A, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights unravel the zymogenic mechanism of the virulence factor gingipain K from Porphyromonas gingivalis, a causative agent of gum disease from the human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
1UNO
 
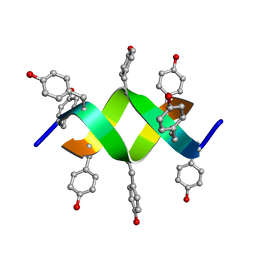 | | Crystal structure of a d,l-alternating peptide | | Descriptor: | H-(L-TYR-D-TYR)4-LYS-OH | | Authors: | Alexopoulos, E, Kuesel, A, Uson, I, Diederichsen, U, Sheldrick, G.M. | | Deposit date: | 2003-09-11 | | Release date: | 2004-09-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Solution and Structure of an Alternating D,L-Peptide
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4IJA
 
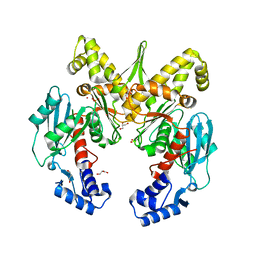 | | Structure of S. aureus methicillin resistance factor MecR2 | | Descriptor: | GLYCEROL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Arede, P, Botelho, T, Guevara, T, Uson, I, Oliveira, D.C, Gomis-Ruth, F.X. | | Deposit date: | 2012-12-21 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Function Studies of the Staphylococcal Methicillin Resistance Antirepressor MecR2.
J.Biol.Chem., 288, 2013
|
|
6F63
 
 | |
1H9I
 
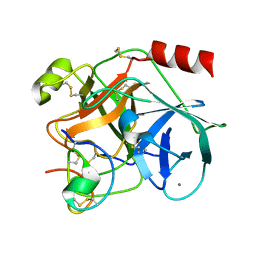 | | COMPLEX OF EETI-II MUTANT WITH PORCINE TRYPSIN | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPSIN INHIBITOR II | | Authors: | Kraetzner, R, Wentzel, A, Kolmar, H, Uson, I. | | Deposit date: | 2001-03-12 | | Release date: | 2004-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Ecballium Elaterium Trypsin Inhibitor II (Eeti-II): A Rigid Molecular Scaffold
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1H9H
 
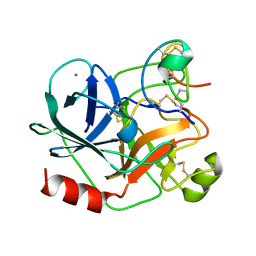 | | COMPLEX OF EETI-II WITH PORCINE TRYPSIN | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPSIN INHIBITOR II | | Authors: | Kraetzner, R, Wentzel, A, Kolmar, H, Uson, I. | | Deposit date: | 2001-03-12 | | Release date: | 2004-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Ecballium Elaterium Trypsin Inhibitor II (Eeti-II): A Rigid Molecular Scaffold
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6GS3
 
 | |
7KFL
 
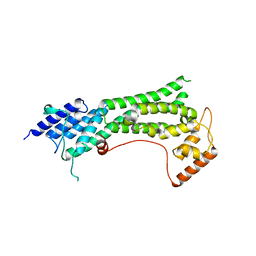 | | Crystal structure of the cargo-binding domain from the plant class XI myosin (MyoXIk) | | Descriptor: | Myosin-17 | | Authors: | Turowski, V.R, Ruiz, D.M, Nascimento, A.F.Z, Millan, C, Sammito, M.D, Juanhuix, J, Cremonesi, A.S, Uson, I, Giuseppe, P.O, Murakami, M.T. | | Deposit date: | 2020-10-14 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the class XI myosin globular tail reveals evolutionary hallmarks for cargo recognition in plants.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7QEC
 
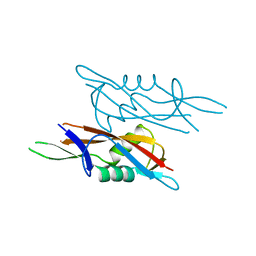 | | Crystal structure of SlpA - domain II, domain that is involved in the self-assembly of the S-layer from Lactobacillus amylovorus | | Descriptor: | S-layer | | Authors: | Eder, M, Dordic, A, Millan, C, Sagmeister, T, Uson, I, Pavkov-Keller, T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QFI
 
 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain I (aa 31-182) | | Descriptor: | CALCIUM ION, SlpX | | Authors: | Sagmeister, T, Damisch, E, Millan, C, Uson, I, Eder, M, Pavkov-Keller, T. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3GWH
 
 | | Crystallographic Ab Initio protein solution far below atomic resolution | | Descriptor: | PHOSPHATE ION, Transcriptional antiterminator (BglG family) | | Authors: | Rodriguez, D.D, Grosse, C, Himmel, S, Gonzalez, C, Becker, S, Sheldrick, G.M, Uson, I. | | Deposit date: | 2009-04-01 | | Release date: | 2010-04-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic ab initio protein structure solution below atomic resolution
Nat.Methods, 6, 2009
|
|
3I1D
 
 | | Distinct recognition of three-way DNA junctions by the two enantiomers of a metallo-supramolecular cylinder ('helicate') | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', FE (II) ION, N-[(1E)-PYRIDIN-2-YLMETHYLENE]-N-[4-(4-{[(1E)-PYRIDIN-2-YLMETHYLENE]AMINO}BENZYL)PHENYL]AMINE | | Authors: | Boer, D.R, Uson, I, Hannon, M.J, Coll, M. | | Deposit date: | 2009-06-26 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Self-Assembly of Functionalizable Two-Component 3D DNA Arrays through the Induced Formation of DNA Three-Way-Junction Branch Points by Supramolecular Cylinders.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
1E1Z
 
 | | Crystal structure of an Arylsulfatase A mutant C69S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION | | Authors: | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | Deposit date: | 2000-05-12 | | Release date: | 2001-05-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis.
J. Mol. Biol., 305, 2001
|
|
1E33
 
 | | Crystal structure of an Arylsulfatase A mutant P426L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION | | Authors: | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | Deposit date: | 2000-06-06 | | Release date: | 2001-05-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Defective oligomerization of arylsulfatase a as a cause of its instability in lysosomes and metachromatic leukodystrophy.
J. Biol. Chem., 277, 2002
|
|
1F94
 
 | | THE 0.97 RESOLUTION STRUCTURE OF BUCANDIN, A NOVEL TOXIN ISOLATED FROM THE MALAYAN KRAIT | | Descriptor: | BUCANDIN | | Authors: | Kuhn, P, Deacon, A.M, Comoso, S, Rajaseger, G, Kini, R.M, Uson, I, Kolatkar, P.R. | | Deposit date: | 2000-07-06 | | Release date: | 2000-07-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The atomic resolution structure of bucandin, a novel toxin isolated from the Malayan krait, determined by direct methods.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1E2S
 
 | | Crystal structure of an Arylsulfatase A mutant C69A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION, ... | | Authors: | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | Deposit date: | 2000-05-24 | | Release date: | 2000-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis.
J. Mol. Biol., 305, 2001
|
|
