3N24
 
 | |
3MN2
 
 | |
3Q94
 
 | | The crystal structure of fructose 1,6-bisphosphate aldolase from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, ACETATE ION, Fructose-bisphosphate aldolase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-07 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | The crystal structure of fructose 1,6-bisphosphate aldolase from Bacillus anthracis str. 'Ames Ancestor'.
To be Published
|
|
3OM8
 
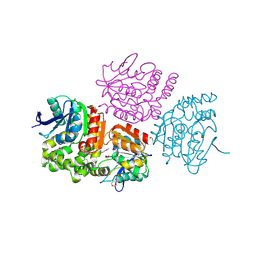 | | The crystal structure of a hydrolase from Pseudomonas aeruginosa PA01 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Probable hydrolase | | Authors: | Tan, K, Chhor, G, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of a hydrolase from Pseudomonas aeruginosa PA01
To be Published
|
|
3ED5
 
 | |
3EDP
 
 | |
3ERM
 
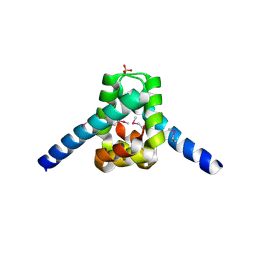 | |
3FC7
 
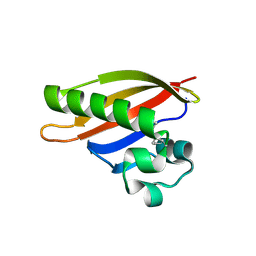 | |
3FG9
 
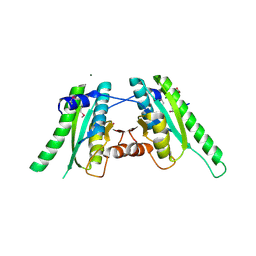 | | The crystal structure of an universal stress protein UspA family protein from Lactobacillus plantarum WCFS1 | | Descriptor: | ACETATE ION, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Tan, K, Li, H, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-05 | | Release date: | 2008-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The crystal structure of an universal stress protein UspA family protein from Lactobacillus plantarum WCFS1
To be Published
|
|
3FK8
 
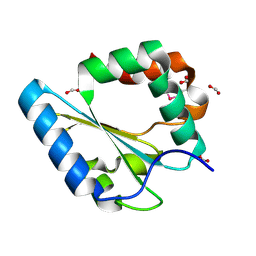 | |
3F1B
 
 | | The crystal structure of a TetR-like transcriptional regulator from Rhodococcus sp. RHA1. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, TetR-like transcriptional regulator | | Authors: | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of a TetR-like transcriptional regulator from Rhodococcus sp. RHA1.
To be Published
|
|
3FFH
 
 | |
3F5B
 
 | | The crystal structure of aminoglycoside N(6')acetyltransferase from Legionella pneumophila subsp. pneumophila str. Philadelphia 1. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aminoglycoside N(6')acetyltransferase, ... | | Authors: | Tan, K, Wu, R, Perez, V, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of aminoglycoside N(6')acetyltransferase from Legionella pneumophila subsp. pneumophila str. Philadelphia 1.
To be Published
|
|
3F5R
 
 | | The crystal structure of a subunit of the heterodimeric FACT complex (Spt16p-Pob3p). | | Descriptor: | CHLORIDE ION, FACT complex subunit POB3, FORMIC ACID, ... | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a subunit of the heterodimeric FACT complex (Spt16p-Pob3p).
To be Published
|
|
3FRM
 
 | | The crystal structure of a functionally unknown conserved protein from Staphylococcus epidermidis ATCC 12228. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SODIUM ION, SULFATE ION, ... | | Authors: | Tan, K, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-08 | | Release date: | 2009-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein from Staphylococcus epidermidis ATCC 12228.
To be Published
|
|
3DSB
 
 | |
6W4B
 
 | | The crystal structure of Nsp9 RNA binding protein of SARS CoV-2 | | Descriptor: | Non-structural protein 9 | | Authors: | Tan, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-10 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of Nsp9 replicase protein of COVID-19
To Be Published
|
|
3FZ4
 
 | | The crystal structure of a possible arsenate reductase from Streptococcus mutans UA159 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Putative arsenate reductase, ... | | Authors: | Tan, K, Hatzos, C, Shackelford, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-10 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The crystal structure of a possible arsenate reductase from Streptococcus mutans UA159.
To be Published
|
|
3DPJ
 
 | | The crystal structure of a TetR transcription regulator from Silicibacter pomeroyi DSS | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Transcription regulator, ... | | Authors: | Tan, K, Li, H, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-08 | | Release date: | 2008-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a TetR transcription regulator from Silicibacter pomeroyi DSS
To be Published
|
|
3E0X
 
 | |
3G74
 
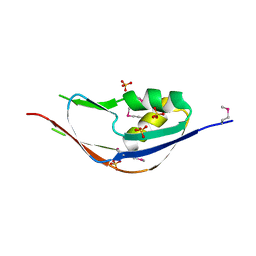 | | Crystal structure of a functionally unknown protein from Eubacterium ventriosum ATCC 27560 | | Descriptor: | Protein of unknown function, SULFATE ION | | Authors: | Tan, K, Sather, A, Marshall, N, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-09 | | Release date: | 2009-03-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The crystal structure of a functionally unknown protein from Eubacterium ventriosum ATCC 27560
To be Published
|
|
3FQ6
 
 | |
3G8W
 
 | | Crystal structure of a probable acetyltransferase from Staphylococcus epidermidis ATCC 12228 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CITRATE ANION, Lactococcal prophage ps3 protein 05 | | Authors: | Tan, K, Sather, A, Marshall, N, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of a probable acetyltransferase from Staphylococcus epidermidis ATCC 12228.
To be Published
|
|
1ZA4
 
 | |
3DNH
 
 | | The crystal structure of the protein Atu2129 (unknown function) from Agrobacterium tumefaciens str. C58 | | Descriptor: | uncharacterized protein Atu2129 | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-02 | | Release date: | 2008-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The crystal structure of the protein Atu2129 (unknown function) from Agrobacterium tumefaciens str. C58
To be Published
|
|
