7N5I
 
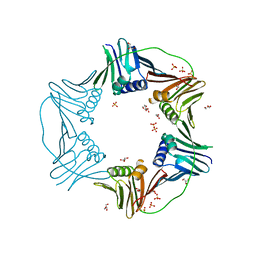 | | PCNA from Thermococcus gammatolerans: crystal I, collection 1, 1.95 A, 5.22 MGy | | Descriptor: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | Authors: | Marin-Tovar, Y, Rudino-Pinera, E. | | Deposit date: | 2021-06-05 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
7N5N
 
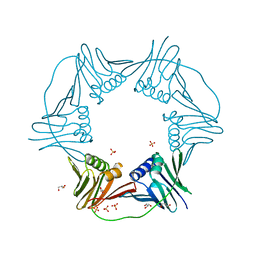 | | PCNA from Thermococcus gammatolerans: crystal III, collection 15, 2.20 A, 28.7 MGy | | Descriptor: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | Authors: | Marin-Tovar, Y, Rudino-Pinera, E. | | Deposit date: | 2021-06-05 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
7N5J
 
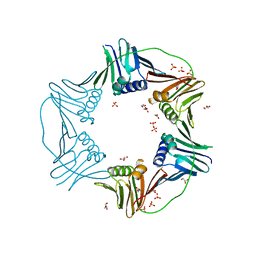 | | PCNA from Thermococcus gammatolerans: crystal I, collection 5, 2.82 A, 89.1 MGy | | Descriptor: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | Authors: | Marin-Tovar, Y, Rudino-Pinera, E. | | Deposit date: | 2021-06-05 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
7N5L
 
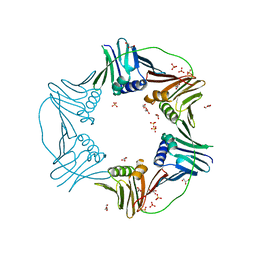 | | PCNA from Thermococcus gammatolerans: crystal II, collection 20, 3.07 A, 77.0 MGy | | Descriptor: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | Authors: | Marin-Tovar, Y, Rudino-Pinera, E. | | Deposit date: | 2021-06-05 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
6PWQ
 
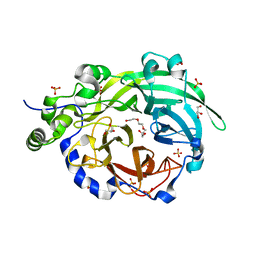 | | Crystal structure of Levansucrase from Bacillus subtilis mutant S164A at 2.6 A | | Descriptor: | CALCIUM ION, GLYCEROL, Glycoside hydrolase family 68 protein, ... | | Authors: | Diaz-Vilchis, A, Rodriguez-Alegria, M.E, Ortiz-Soto, M.E, Rudino-Pinera, E, Lopez-Munguia, A. | | Deposit date: | 2019-07-23 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Implications of the mutation S164A on Bacillus subtilis levansucrase product specificity and insights into protein interactions acting upon levan synthesis.
Int.J.Biol.Macromol., 161, 2020
|
|
6Q29
 
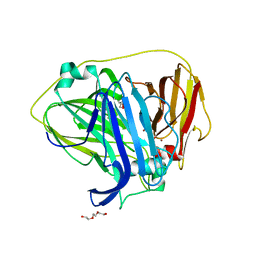 | |
2WME
 
 | | Crystallographic structure of betaine aldehyde dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | BETA-MERCAPTOETHANOL, BETAINE ALDEHYDE DEHYDROGENASE, GLYCEROL, ... | | Authors: | Gonzalez-Segura, L, Rudino-Pinera, E, Munoz-Clares, R.A, Horjales, E. | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of a Ternary Complex of Betaine Aldehyde Dehydrogenase from Pseudomonas Aeruginosa Provides New Insight Into the Reaction Mechanism and Shows a Novel Binding Mode of the 2'- Phosphate of Nadp(+) and a Novel Cation Binding Site.
J.Mol.Biol., 385, 2009
|
|
5EYW
 
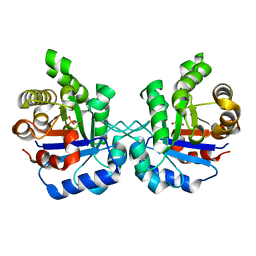 | | Crystal structure of Litopenaeus vannamei triosephosphate isomerase complexed with 2-Phosphoglycolic acid | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Carrasco-Miranda, J.S, Lopez-Zavala, A.A, Brieba, L.G, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2015-11-25 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights from a novel invertebrate triosephosphate isomerase from Litopenaeus vannamei.
Biochim.Biophys.Acta, 1864, 2016
|
|
5FNX
 
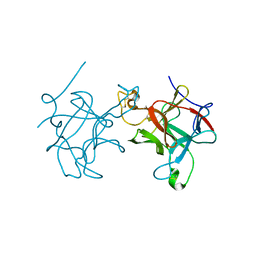 | |
5FZY
 
 | |
5G00
 
 | |
5FNW
 
 | |
5FZZ
 
 | |
5G31
 
 | |
2VDT
 
 | | Crystallographic structure of Levansucrase from Bacillus subtilis mutant S164A | | Descriptor: | CALCIUM ION, LEVANSUCRASE | | Authors: | Ortiz-Soto, M.E, Rivera, M, Rudino-Pinera, E, Olvera, C, Lopez-Munguia, A. | | Deposit date: | 2007-10-11 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Selected Mutations in Bacillus Subtilis Levansucrase Semi-Conserved Regions Affecting its Biochemical Properties
Protein Eng.Des.Sel., 21, 2008
|
|
2WOX
 
 | | Betaine aldehyde dehydrogenase from Pseudomonas aeruginosa with NAD(P) H-catalytic thiol adduct. | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, BETA-MERCAPTOETHANOL, BETAINE ALDEHYDE DEHYDROGENASE, ... | | Authors: | Gonzalez-Segura, L, Rudino-Pinera, E, Diaz-Sanchez, A.G, Munoz-Clares, R.A. | | Deposit date: | 2009-07-30 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel NADPH-cysteine covalent adduct found in the active site of an aldehyde dehydrogenase.
Biochem. J., 439, 2011
|
|
2Y70
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF GM23, MUTANT G89D, AN EXAMPLE OF CATALYTIC MIGRATION FROM TIM TO THIAMIN PHOSPHATE SYNTHASE. | | Descriptor: | ACETATE ION, SULFATE ION, TRIOSE-PHOSPHATE ISOMERASE | | Authors: | Saab-Rincon, G, Olvera, L, Olvera, M, Rudino-Pinera, E, Soberon, X, Morett, E. | | Deposit date: | 2011-01-27 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolutionary Walk between (Beta/Alpha)(8) Barrels: Catalytic Migration from Triosephosphate Isomerase to Thiamin Phosphate Synthase.
J.Mol.Biol., 416, 2012
|
|
2Y6Z
 
 | | Crystallographic structure of GM23 an example of Catalytic migration from TIM to thiamin phosphate synthase. | | Descriptor: | PYROPHOSPHATE 2-, THIAMIN PHOSPHATE, TRIOSE-PHOSPHATE ISOMERASE | | Authors: | Saab-Rincon, G, Olvera, L, Olvera, M, Rudino-Pinera, E, Soberon, X, Morett, E. | | Deposit date: | 2011-01-27 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolutionary Walk between (Beta/Alpha)(8) Barrels: Catalytic Migration from Triosephosphate Isomerase to Thiamin Phosphate Synthase.
J.Mol.Biol., 416, 2012
|
|
4A2E
 
 | | Crystal Structure of a Coriolopsis gallica Laccase at 1.7 A Resolution pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, LACCASE | | Authors: | De La Mora, E, Valderrama, B, Horjales, E, Rudino-Pinera, E. | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Changes Caused by Radiation-Induced Reduction and Radiolysis: The Effect of X-Ray Absorbed Dose in a Fungal Multicopper Oxidase
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4A2H
 
 | | Crystal Structure of Laccase from Coriolopsis gallica pH 7.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | De La Mora, E, Valderrama, B, Horjales, E, Rudino-Pinera, E. | | Deposit date: | 2011-09-27 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Changes Caused by Radiation-Induced Reduction and Radiolysis: The Effect of X-Ray Absorbed Dose in a Fungal Multicopper Oxidase
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4A2D
 
 | | Coriolopsis gallica Laccase T2 Copper Depleted at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, LACCASE, ... | | Authors: | De La Mora, E, Valderrama, B, Horjales, E, Rudino-Pinera, E. | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Changes Caused by Radiation-Induced Reduction and Radiolysis: The Effect of X-Ray Absorbed Dose in a Fungal Multicopper Oxidase
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4A2F
 
 | | Coriolopsis gallica laccase collected at 12.65 keV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, LACCASE | | Authors: | De la Mora, E, Rudino-Pinera, E. | | Deposit date: | 2011-09-26 | | Release date: | 2012-04-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Changes Caused by Radiation-Induced Reduction and Radiolysis: The Effect of X-Ray Absorbed Dose in a Fungal Multicopper Oxidase
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4A2G
 
 | | Coriolopsis gallica laccase collected at 8.98 keV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, LACCASE | | Authors: | De la Mora, E, Rudino-Pinera, E. | | Deposit date: | 2011-09-26 | | Release date: | 2012-04-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Changes Caused by Radiation-Induced Reduction and Radiolysis: The Effect of X-Ray Absorbed Dose in a Fungal Multicopper Oxidase
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3ZJ5
 
 | | NEUROSPORA CRASSA CATALASE-3 EXPRESSED IN E. COLI, ORTHORHOMBIC FORM. | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-ETHOXYETHOXY)ETHANOL, CATALASE-3, ... | | Authors: | Zarate-Romero, A, Rudino-Pinera, E. | | Deposit date: | 2013-01-17 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Conformational Stability and Crystal Packing: Polymorphism in Neurospora Crassa Cat-3
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3ZJ4
 
 | |
