2QTW
 
 | | The Crystal Structure of PCSK9 at 1.9 Angstroms Resolution Reveals structural homology to Resistin within the C-terminal domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Hampton, E.N, Knuth, M.W, Li, J, Harris, J.L, Lesley, S.A, Spraggon, G. | | Deposit date: | 2007-08-02 | | Release date: | 2007-09-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The self-inhibited structure of full-length PCSK9 at 1.9 A reveals structural homology with resistin within the C-terminal domain.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PIS
 
 | | Efforts toward Expansion of the Genetic Alphabet: Structure and Replication of Unnatural Base Pairs | | Descriptor: | DNA (5'-D(*CP*GP*(CBR)P*GP*AP*AP*(FFD)P*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Matsuda, S, Fillo, J.D, Henry, A.A, Wilkins, S.J, Rai, P, Dwyer, T.J, Geierstanger, B.H, Wemmer, D.E, Schultz, P.G, Spraggon, G, Romesberg, F.E. | | Deposit date: | 2007-04-13 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Efforts toward expansion of the genetic alphabet: structure and replication of unnatural base pairs.
J.Am.Chem.Soc., 129, 2007
|
|
2Q7D
 
 | | Crystal Structure of Human Inositol 1,3,4-Trisphosphate 5/6-kinase (ITPK1) in complex with AMPPNP and Mn2+ | | Descriptor: | Inositol-tetrakisphosphate 1-kinase, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chamberlain, P.P, Lesley, S.A, Spraggon, G. | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Integration of inositol phosphate signaling pathways via human ITPK1.
J.Biol.Chem., 282, 2007
|
|
2V9K
 
 | | Crystal structure of human PUS10, a novel pseudouridine synthase. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | McCleverty, C.J, Hornsby, M, Spraggon, G, Kreusch, A. | | Deposit date: | 2007-08-23 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Pus10, a Novel Pseudouridine Synthase.
J.Mol.Biol., 373, 2007
|
|
4AE0
 
 | |
2AG6
 
 | | Crystal structure of p-bromo-l-phenylalanine-tRNA sythetase in complex with p-bromo-l-phenylalanine | | Descriptor: | 4-BROMO-L-PHENYLALANINE, Tyrosyl-tRNA synthetase | | Authors: | Turner, J.M, Graziano, J, Spraggon, G, Schultz, P.G. | | Deposit date: | 2005-07-26 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural plasticity of an aminoacyl-tRNA synthetase active site
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2AQX
 
 | | Crystal Structure of the Catalytic and CaM-Binding domains of Inositol 1,4,5-Trisphosphate 3-Kinase B | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PREDICTED: inositol 1,4,5-trisphosphate 3-kinase B | | Authors: | Chamberlain, P.P, Sandberg, M.L, Sauer, K, Cooke, M.P, Lesley, S.A, Spraggon, G. | | Deposit date: | 2005-08-18 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into enzyme regulation for inositol 1,4,5-trisphosphate 3-kinase B
Biochemistry, 44, 2005
|
|
4AE1
 
 | |
1ZH6
 
 | | Crystal Structure of p-acetylphenylalanine-tRNA synthetase in complex with p-acetylphenylalanine | | Descriptor: | 4-ACETYL-L-PHENYLALANINE, BETA-MERCAPTOETHANOL, Tyrosyl-tRNA synthetase | | Authors: | Turner, J.M, Graziano, J, Spraggon, G, Schultz, P.G. | | Deposit date: | 2005-04-22 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural characterization of a p-acetylphenylalanyl aminoacyl-tRNA synthetase.
J.Am.Chem.Soc., 127, 2005
|
|
1ZLT
 
 | | Crystal Structure of Chk1 Complexed with a Hymenaldisine Analog | | Descriptor: | (4Z)-4-(2-AMINO-5-OXO-3,5-DIHYDRO-4H-IMIDAZOL-4-YLIDENE)-2,3-DICHLORO-4,5,6,7-TETRAHYDROPYRROLO[2,3-C]AZEPIN-8(1H)-ONE, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Lee, C.C, Ng, K, Wan, Y, Gray, N, Spraggon, G. | | Deposit date: | 2005-05-09 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of Chk1 Complexed with a Hymenaldisine Analog
To be Published
|
|
1ZH0
 
 | | Crystal Structure of L-3-(2-napthyl)alanine-tRNA synthetase in complex with L-3-(2-napthyl)alanine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-(2-NAPHTHYL)-ALANINE, Tyrosyl-tRNA synthetase | | Authors: | Turner, J.M, Graziano, J, Spraggon, G, Schultz, P.G. | | Deposit date: | 2005-04-22 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural plasticity of an aminoacyl-tRNA synthetase active site
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3QE4
 
 | | An evolved aminoacyl-tRNA Synthetase with atypical polysubstrate specificity | | Descriptor: | 4-cyano-L-phenylalanine, Tyrosyl-tRNA synthetase | | Authors: | Young, D.D, Young, T.S, Jahnz, M, Ahmad, I, Spraggon, G, Schultz, P.G. | | Deposit date: | 2011-01-19 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Evolved Aminoacyl-tRNA Synthetase with Atypical Polysubstrate Specificity .
Biochemistry, 50, 2011
|
|
3R5L
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayyar, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-18 | | Release date: | 2012-01-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5R
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | Descriptor: | COENZYME F420, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
2H7J
 
 | | Crystal Structure of Cathepsin S in complex with a Nonpeptidic Inhibitor. | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, Cathepsin S, N-[(1S)-1-{1-[(1R,3E)-1-ACETYLPENT-3-EN-1-YL]-1H-1,2,3-TRIAZOL-4-YL}-1,2-DIMETHYLPROPYL]BENZAMIDE | | Authors: | Patterson, A.W, Wood, W.J, Hornsby, M, Lesley, S, Spraggon, G, Ellman, J.A. | | Deposit date: | 2006-06-02 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of selective, nonpeptidic nitrile inhibitors of cathepsin s using the substrate activity screening method.
J.Med.Chem., 49, 2006
|
|
3R5Z
 
 | | Structure of a Deazaflavin-dependent reductase from Nocardia farcinica, with co-factor F420 | | Descriptor: | COENZYME F420, Putative uncharacterized protein, SULFATE ION | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5P
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | Descriptor: | Deazaflavin-dependent nitroreductase, SULFATE ION | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5Y
 
 | | Structure of a Deazaflavin-dependent nitroreductase from Nocardia farcinica, with co-factor F420 | | Descriptor: | COENZYME F420, Putative uncharacterized protein | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5W
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | Descriptor: | COENZYME F420, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
2HXZ
 
 | | Crystal Structure of Cathepsin S in complex with a Nonpeptidic Inhibitor (Hexagonal spacegroup) | | Descriptor: | Cathepsin S, N-[(1S)-1-{1-[(1R,3E)-1-ACETYLPENT-3-EN-1-YL]-1H-1,2,3-TRIAZOL-4-YL}-1,2-DIMETHYLPROPYL]BENZAMIDE, SULFATE ION | | Authors: | Patterson, A.W, Wood, W.J, Hornsby, M, Lesley, S, Spraggon, G, Ellman, J.A. | | Deposit date: | 2006-08-04 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of selective, nonpeptidic nitrile inhibitors of cathepsin s using the substrate activity screening method.
J.Med.Chem., 49, 2006
|
|
1J7J
 
 | |
1FZE
 
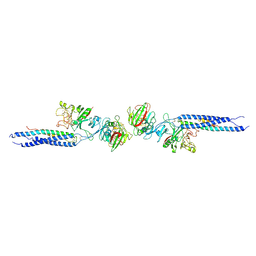 | | CRYSTAL STRUCTURE OF FRAGMENT DOUBLE-D FROM HUMAN FIBRIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FIBRINOGEN | | Authors: | Everse, S.J, Spraggon, G, Veerapandian, L, Doolittle, R.F. | | Deposit date: | 1998-12-23 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformational changes in fragments D and double-D from human fibrin(ogen) upon binding the peptide ligand Gly-His-Arg-Pro-amide.
Biochemistry, 38, 1999
|
|
1FZF
 
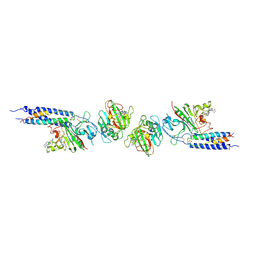 | | CRYSTAL STRUCTURE OF FRAGMENT DOUBLE-D FROM HUMAN FIBRIN WITH THE PEPTIDE LIGAND GLY-HIS-ARG-PRO-AMIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FIBRINOGEN | | Authors: | Everse, S.J, Spraggon, G, Veerapandian, L, Doolittle, R.F. | | Deposit date: | 1998-12-28 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational changes in fragments D and double-D from human fibrin(ogen) upon binding the peptide ligand Gly-His-Arg-Pro-amide.
Biochemistry, 38, 1999
|
|
1FZG
 
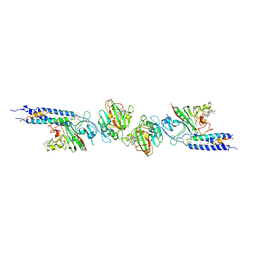 | | CRYSTAL STRUCTURE OF FRAGMENT D FROM HUMAN FIBRINOGEN WITH THE PEPTIDE LIGAND GLY-HIS-ARG-PRO-AMIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FIBRINOGEN | | Authors: | Everse, S.J, Spraggon, G, Veerapandian, L, Doolittle, R.F. | | Deposit date: | 1999-01-01 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational changes in fragments D and double-D from human fibrin(ogen) upon binding the peptide ligand Gly-His-Arg-Pro-amide.
Biochemistry, 38, 1999
|
|
1LWU
 
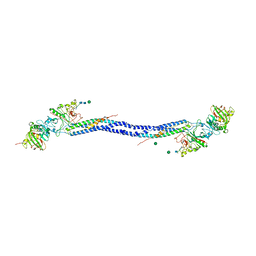 | | Crystal structure of fragment D from lamprey fibrinogen complexed with the peptide Gly-His-Arg-Pro-amide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, Z, Spraggon, G, Pandi, L, Everse, S.J, Riley, M, Doolittle, R.F. | | Deposit date: | 2002-06-03 | | Release date: | 2002-08-23 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of fragment D from lamprey fibrinogen complexed with the peptide Gly-His-Arg-Pro-amide.
Biochemistry, 41, 2002
|
|
