7MHO
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 298 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
7MNG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor VBY-825 (Partial Occupancy) | | Descriptor: | (2R,3S)-N-cyclopropyl-3-{[(2R)-3-(cyclopropylmethanesulfonyl)-2-{[(1S)-2,2,2-trifluoro-1-(4-fluorophenyl)ethyl]amino}propanoyl]amino}-2-hydroxypentanamide (non-preferred name), 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2021-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7MHI
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 298 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHQ
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 310 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9601 Å) | | Cite: | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
7MHH
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 277 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1908 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHP
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 298 K at high humidity | | Descriptor: | 3C-like proteinase, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.0005 Å) | | Cite: | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
7MRR
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Leupeptin | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, LEUPEPTIN | | Authors: | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2021-05-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
2CE4
 
 | | Manganese Superoxide Dismutase (Mn-SOD) from Deinococcus radiodurans | | Descriptor: | MANGANESE (II) ION, SUPEROXIDE DISMUTASE [MN] | | Authors: | Dennis, R, Micossi, E, McCarthy, J, Moe, E, Gordon, E, Leonard, G, McSweeney, S. | | Deposit date: | 2006-02-03 | | Release date: | 2006-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the manganese superoxide dismutase from Deinococcus radiodurans in two crystal forms.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 62, 2006
|
|
2CDY
 
 | | Manganese Superoxide Dismutase (Mn-SOD) from Deinococcus radiodurans | | Descriptor: | MANGANESE (II) ION, SUPEROXIDE DISMUTASE [MN] | | Authors: | Dennis, R, Micossi, E, McCarthy, J, Moe, E, Gordon, E, Leonard, G, McSweeney, S. | | Deposit date: | 2006-01-31 | | Release date: | 2006-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the manganese superoxide dismutase from Deinococcus radiodurans in two crystal forms.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 62, 2006
|
|
1ZIQ
 
 | | Deuterated gammaE crystallin in D2O solvent | | Descriptor: | ACETATE ION, Gamma crystallin E | | Authors: | Artero, J.B, Hartlein, M, McSweeney, S, Timmins, P. | | Deposit date: | 2005-04-27 | | Release date: | 2005-11-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A comparison of refined X-ray structures of hydrogenated and perdeuterated rat gammaE-crystallin in H2O and D2O.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZGT
 
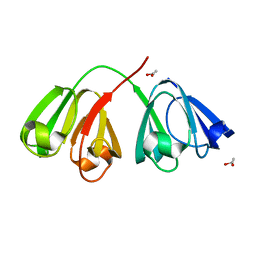 | | Structure of hydrogenated rat gamma E crystallin in H2O | | Descriptor: | ACETATE ION, Gamma crystallin E | | Authors: | Artero, J.B, Hartlein, M, McSweeney, S, Timmins, P. | | Deposit date: | 2005-04-22 | | Release date: | 2005-11-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A comparison of refined X-ray structures of hydrogenated and perdeuterated rat gammaE-crystallin in H2O and D2O.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZIR
 
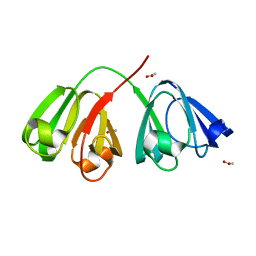 | | Deuterated gammaE crystallin in H2O solvent | | Descriptor: | ACETATE ION, Gamma crystallin E | | Authors: | Artero, J.B, Hartlein, M, McSweeney, S, Timmins, P. | | Deposit date: | 2005-04-27 | | Release date: | 2005-11-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A comparison of refined X-ray structures of hydrogenated and perdeuterated rat gammaE-crystallin in H2O and D2O.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2C2F
 
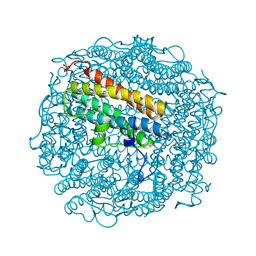 | | Dps from Deinococcus radiodurans | | Descriptor: | DNA-BINDING STRESS RESPONSE PROTEIN, FE (III) ION, GLYCEROL, ... | | Authors: | Romao, C.V, Mitchell, E, McSweeney, S. | | Deposit date: | 2005-09-28 | | Release date: | 2006-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The Crystal Structure of Deinococcus Radiodurans Dps Protein (Dr2263) Reveals the Presence of a Novel Metal Centre in the N Terminus.
J.Biol.Inorg.Chem., 11, 2006
|
|
2BHY
 
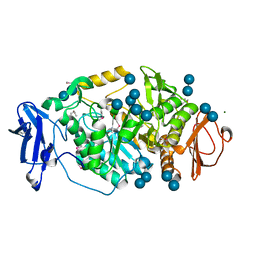 | | Crystal structure of Deinococcus radiodurans maltooligosyltrehalose trehalohydrolase in complex with trehalose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | Timmins, J, Leiros, H.-K.S, Leonard, G, Leiros, I, McSweeney, S. | | Deposit date: | 2005-01-20 | | Release date: | 2005-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Maltooligosyltrehalose Trehalohydrolase from Deinococcus Radiodurans in Complex with Disaccharides
J.Mol.Biol., 347, 2005
|
|
2BHU
 
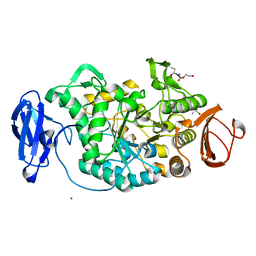 | | Crystal structure of Deinococcus radiodurans maltooligosyltrehalose trehalohydrolase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, MALTOOLIGOSYLTREHALOSE TREHALOHYDROLASE, ... | | Authors: | Timmins, J, Leiros, H.-K.S, Leonard, G, Leiros, I, McSweeney, S. | | Deposit date: | 2005-01-18 | | Release date: | 2005-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of Maltooligosyltrehalose Trehalohydrolase from Deinococcus Radiodurans in Complex with Disaccharides
J.Mol.Biol., 347, 2005
|
|
2BHZ
 
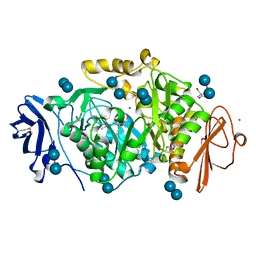 | | Crystal structure of Deinococcus radiodurans maltooligosyltrehalose trehalohydrolase in complex with maltose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | Timmins, J, Leiros, H.-K.S, Leonard, G, Leiros, I, McSweeney, S. | | Deposit date: | 2005-01-20 | | Release date: | 2005-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of Maltooligosyltrehalose Trehalohydrolase from Deinococcus Radiodurans in Complex with Disaccharides
J.Mol.Biol., 347, 2005
|
|
2BOO
 
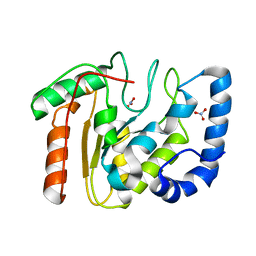 | | The crystal structure of Uracil-DNA N-Glycosylase (UNG) from Deinococcus radiodurans. | | Descriptor: | NITRATE ION, URACIL-DNA GLYCOSYLASE | | Authors: | Leiros, I, Moe, E, Smalas, A.O, McSweeney, S. | | Deposit date: | 2005-04-13 | | Release date: | 2005-07-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Uracil-DNA N-Glycosylase (Ung) from Deinococcus Radiodurans.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2C2U
 
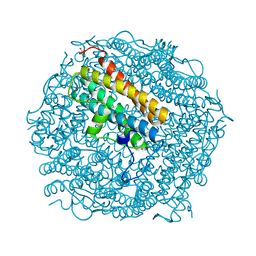 | | Dps from Deinococcus radiodurans | | Descriptor: | DNA-BINDING STRESS RESPONSE PROTEIN, FE (III) ION, SULFATE ION, ... | | Authors: | Romao, C.V, Mitchell, E, McSweeney, S. | | Deposit date: | 2005-09-30 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Crystal Structure of Deinococcus Radiodurans Dps Protein (Dr2263) Reveals the Presence of a Novel Metal Centre in the N Terminus.
J.Biol.Inorg.Chem., 11, 2006
|
|
2C2P
 
 | | The crystal structure of mismatch specific uracil-DNA glycosylase (MUG) from Deinococcus radiodurans | | Descriptor: | ACETATE ION, G/U MISMATCH-SPECIFIC DNA GLYCOSYLASE | | Authors: | Moe, E, Leiros, I, Smalas, A.O, McSweeney, S. | | Deposit date: | 2005-09-29 | | Release date: | 2005-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Crystal Structure of Mismatch Specific Uracil-DNA Glycosylase (Mug) from Deinococcus Radiodurans Reveals a Novel Catalytic Residue and Broad Substrate Specificity
J.Biol.Chem., 281, 2006
|
|
2C2Q
 
 | | The crystal structure of mismatch specific uracil-DNA glycosylase (MUG) from Deinococcus radiodurans. Inactive mutant Asp93Ala. | | Descriptor: | ACETATE ION, G/U MISMATCH-SPECIFIC DNA GLYCOSYLASE | | Authors: | Moe, E, Leiros, I, Smalas, A.O, McSweeney, S. | | Deposit date: | 2005-09-29 | | Release date: | 2005-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Mismatch Specific Uracil-DNA Glycosylase (Mug) from Deinococcus Radiodurans Reveals a Novel Catalytic Residue and Broad Substrate Specificity
J.Biol.Chem., 281, 2006
|
|
1ZIE
 
 | | Hydrogenated gammaE crystallin in D2O solvent | | Descriptor: | ACETATE ION, Gamma crystallin E | | Authors: | Artero, J.B, Hartlein, M, McSweeney, S, Timmins, P. | | Deposit date: | 2005-04-27 | | Release date: | 2005-11-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | A comparison of refined X-ray structures of hydrogenated and perdeuterated rat gammaE-crystallin in H2O and D2O.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6Y8G
 
 | | selenomethionine derivative of ferulic acid esterase (FAE) | | Descriptor: | CADMIUM ION, Endo-1,4-beta-xylanase Y, GLYCEROL | | Authors: | von Stetten, D, Mueller-Dieckmann, C, Carpentier, P, Flot, D. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-20 | | Last modified: | 2023-03-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ID30A-3 (MASSIF-3) - a beamline for macromolecular crystallography at the ESRF with a small intense beam.
J.Synchrotron Radiat., 27, 2020
|
|
4YTA
 
 | | BOND LENGTH ANALYSIS OF ASP, GLU AND HIS RESIDUES IN TRYPSIN AT 1.2A RESOLUTION | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Fisher, S.J, Helliwell, J.R, Blakeley, M.P, Cianci, M, McSweeny, S. | | Deposit date: | 2015-03-17 | | Release date: | 2015-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protonation-state determination in proteins using high-resolution X-ray crystallography: effects of resolution and completeness.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
3UNX
 
 | | Bond length analysis of asp, glu and his residues in subtilisin Carlsberg at 1.26A resolution | | Descriptor: | CALCIUM ION, GLYCEROL, SODIUM ION, ... | | Authors: | Fisher, S.J, Helliwell, J.R, Blakeley, M.P, Cianci, M, McSweeny, S. | | Deposit date: | 2011-11-16 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Protonation-state determination in proteins using high-resolution X-ray crystallography: effects of resolution and completeness.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8G83
 
 | | Structure of NAD+ consuming protein Acinetobacter baumannii TIR domain | | Descriptor: | NAD(+) hydrolase AbTIR | | Authors: | Klontz, E.H, Wang, Y, Glendening, G, Carr, J, Tsibouris, T, Buddula, S, Nallar, S, Soares, A, Snyder, G.A. | | Deposit date: | 2023-02-17 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | The structure of NAD + consuming protein Acinetobacter baumannii TIR domain shows unique kinetics and conformations.
J.Biol.Chem., 299, 2023
|
|
