5WOE
 
 | |
4P2A
 
 | | Structure of mouse VPS26A bound to rat SNX27 PDZ domain | | Descriptor: | MERCURY (II) ION, Sorting nexin-27, Vacuolar protein sorting-associated protein 26A | | Authors: | Clairfeuille, T, Gallon, M, Mas, C, Ghai, R, Teasdale, R, Cullen, P, Collins, B. | | Deposit date: | 2014-03-03 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A unique PDZ domain and arrestin-like fold interaction reveals mechanistic details of endocytic recycling by SNX27-retromer.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7RN0
 
 | |
7RN1
 
 | |
7VHP
 
 | | Structural insights into the membrane microdomain organization by SPFH family proteins | | Descriptor: | ATP-dependent zinc metalloprotease FtsH, Modulator of FtsH protease HflC, Protein HflK | | Authors: | Ma, C.Y, Wang, C.K, Luo, D.Y, Yan, L, Yang, W.X, Li, N.N, Gao, N. | | Deposit date: | 2021-09-22 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into the membrane microdomain organization by SPFH family proteins.
Cell Res., 32, 2022
|
|
7VHQ
 
 | | Structural insights into the membrane microdomain organization by SPFH family proteins | | Descriptor: | ATP-dependent zinc metalloprotease FtsH, Modulator of FtsH protease HflC, Protein HflK | | Authors: | Ma, C.Y, Wang, C.K, Luo, D.Y, Yan, L, Yang, W.X, Li, N.N, Gao, N. | | Deposit date: | 2021-09-22 | | Release date: | 2022-01-19 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into the membrane microdomain organization by SPFH family proteins.
Cell Res., 32, 2022
|
|
7KX5
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with noncovalent inhibitor Jun8-76-3A | | Descriptor: | 3C-like proteinase, GLYCEROL, N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]furan-2-carboxamide | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Di- and Trihaloacetamides as Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity.
J.Am.Chem.Soc., 143, 2021
|
|
2MT4
 
 | |
6ITC
 
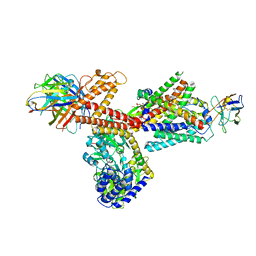 | | Structure of a substrate engaged SecA-SecY protein translocation machine | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Ma, C.Y, Wu, X.F, Sun, D.J, Park, E.Y, Rapoport, T.A, Gao, N, Long, L. | | Deposit date: | 2018-11-21 | | Release date: | 2019-06-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of the substrate-engaged SecA-SecY protein translocation machine.
Nat Commun, 10, 2019
|
|
7YKT
 
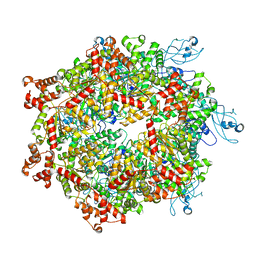 | | Cryo-EM structure of Drg1 hexamer in helical state treated with ADP/AMPPNP/benzo-diazaborine | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase family gene 2 protein | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2022-07-23 | | Release date: | 2022-12-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7YKL
 
 | | Cryo-EM structure of Drg1 hexamer treated with AMPPNP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase family gene 2 protein | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2022-07-22 | | Release date: | 2022-12-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7YKK
 
 | | Cryo-EM structure of Drg1 hexamer treated with ADP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase family gene 2 protein | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2022-07-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7YKZ
 
 | | Cryo-EM structure of Drg1 hexamer in the planar state treated with ADP/AMPPNP/Diazaborine | | Descriptor: | 2-(TOLUENE-4-SULFONYL)-2H-BENZO[D][1,2,3]DIAZABORININ-1-OL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2022-07-25 | | Release date: | 2022-12-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7WD3
 
 | | Cryo-EM structure of Drg1 hexamer treated with ATP and benzo-diazaborine | | Descriptor: | 2-(TOLUENE-4-SULFONYL)-2H-BENZO[D][1,2,3]DIAZABORININ-1-OL, ADENOSINE-5'-TRIPHOSPHATE, ATPase family gene 2 protein | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7WBB
 
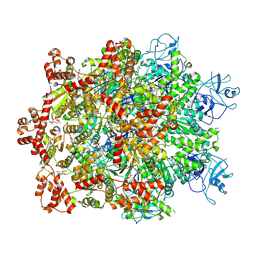 | | Cryo-EM structure of substrate engaged Drg1 hexamer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AFG2 isoform 1, substrate | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2021-12-16 | | Release date: | 2022-12-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
2LY7
 
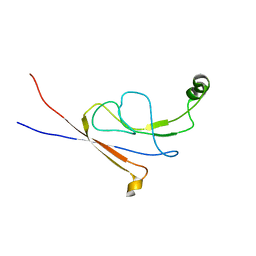 | | B-flap domain of RNA polymerase (B. subtilis) | | Descriptor: | DNA-directed RNA polymerase subunit beta | | Authors: | Mobli, M. | | Deposit date: | 2012-09-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RNA polymerase-induced remodelling of NusA produces a pause enhancement complex.
Nucleic Acids Res., 43, 2015
|
|
4P2J
 
 | |
4P2I
 
 | | Crystal structure of the mouse SNX19 PX domain | | Descriptor: | MKIAA0254 protein | | Authors: | Collins, B.M. | | Deposit date: | 2014-03-04 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Different Phosphoinositide Specificities of the PX Domains of Sorting Nexins Regulating G-protein Signaling.
J.Biol.Chem., 289, 2014
|
|
5XM7
 
 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-((2S,3R)-3-amino-2-hydroxy-4-phenylbutanamido)-N-hydroxy-4-methylpentanamide | | Descriptor: | (2S)-4-methyl-N-[(1R)-2-(oxidanylamino)-2-oxidanylidene-1-phenyl-ethyl]-2-[(phenylmethyl)carbamoylamino]pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-05-12 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Development of peptidomimetic hydroxamates as PfA-M1 and PfA-M17 dual inhibitors: Biological evaluation and structural characterization by cocrystallization
Chin.Chem.Lett., 33, 2022
|
|
5Y1T
 
 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,3-dimethylbenzyl)ureido)-N-hydroxy-4-methylpentanamide | | Descriptor: | (2S)-2-[(2,3-dimethylphenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Development of peptidomimetic hydroxamates as PfA-M1 and PfA-M17 dual inhibitors: Biological evaluation and structural characterization by cocrystallization
Chin.Chem.Lett., 33, 2022
|
|
6LCY
 
 | | Crystal structure of Synaptotagmin-7 C2B in complex with IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Synaptotagmin-7 | | Authors: | Zhang, Y, Zhang, X, Rao, F, Wang, C. | | Deposit date: | 2019-11-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 5-IP 7 is a GPCR messenger mediating neural control of synaptotagmin-dependent insulin exocytosis and glucose homeostasis.
Nat Metab, 3, 2021
|
|
6ZDH
 
 | | SARS-CoV-2 Spike glycoprotein in complex with a neutralizing antibody EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5UF7
 
 | | CRYSTAL STRUCTURE OF MUNC13-1 MUN DOMAIN | | Descriptor: | Protein unc-13 homolog A | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2017-01-03 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
5UE8
 
 | | The crystal structure of Munc13-1 C1C2BMUN domain | | Descriptor: | CHLORIDE ION, Protein unc-13 homolog A, ZINC ION | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
9J1W
 
 | |
