1RM4
 
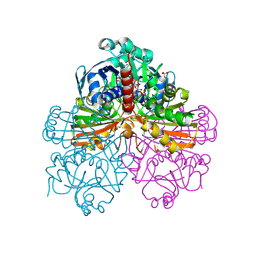 | | Crystal structure of recombinant photosynthetic glyceraldehyde-3-phosphate dehydrogenase A4 isoform, complexed with NADP | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase A, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Sparla, F, Fermani, S, Falini, G, Ripamonti, A, Sabatino, P, Pupillo, P, Trost, P. | | Deposit date: | 2003-11-27 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coenzyme Site-directed Mutants of Photosynthetic A(4)-GAPDH Show Selectively Reduced NADPH-dependent Catalysis, Similar to Regulatory AB-GAPDH Inhibited by Oxidized Thioredoxin
J.Mol.Biol., 340, 2004
|
|
1RM3
 
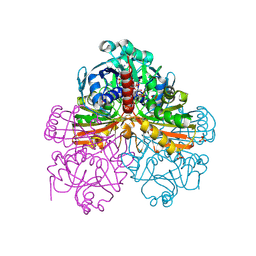 | | Crystal structure of mutant T33A of photosynthetic glyceraldehyde-3-phosphate dehydrogenase A4 isoform, complexed with NADP | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase A, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Sparla, F, Fermani, S, Falini, G, Ripamonti, A, Sabatino, P, Pupillo, P, Trost, P. | | Deposit date: | 2003-11-27 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Coenzyme Site-directed Mutants of Photosynthetic A(4)-GAPDH Show Selectively Reduced NADPH-dependent Catalysis, Similar to Regulatory AB-GAPDH Inhibited by Oxidized Thioredoxin
J.Mol.Biol., 340, 2004
|
|
1RM5
 
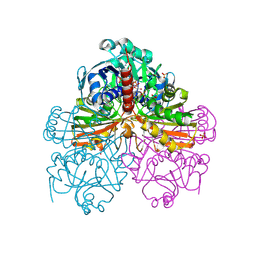 | | Crystal structure of mutant S188A of photosynthetic glyceraldehyde-3-phosphate dehydrogenase A4 isoform, complexed with NADP | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase A, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Sparla, F, Fermani, S, Falini, G, Ripamonti, A, Sabatino, P, Pupillo, P, Trost, P. | | Deposit date: | 2003-11-27 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Coenzyme Site-directed Mutants of Photosynthetic A(4)-GAPDH Show Selectively Reduced NADPH-dependent Catalysis, Similar to Regulatory AB-GAPDH Inhibited by Oxidized Thioredoxin
J.Mol.Biol., 340, 2004
|
|
3CTK
 
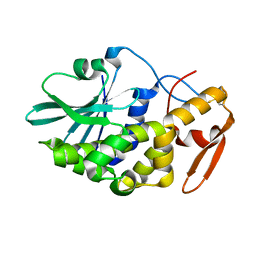 | | Crystal structure of the type 1 RIP bouganin | | Descriptor: | rRNA N-glycosidase | | Authors: | Fermani, S, Tosi, G, Falini, G, Ripamonti, A, Farini, V, Bolognesi, A, Polito, L. | | Deposit date: | 2008-04-14 | | Release date: | 2008-05-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure/function studies on two type 1 ribosome inactivating proteins: Bouganin and lychnin.
J.Struct.Biol., 168, 2009
|
|
7RQ8
 
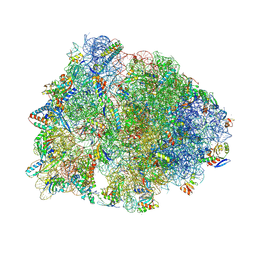 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAs, and aminoacylated P-site tRNA at 2.50A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mitcheltree, M.J, Pisipati, A, Syroegin, E.A, Silvestre, K.J, Klepacki, D, Mason, J.D, Terwilliger, D.W, Testolin, G, Pote, A.R, Wu, K.J.Y, Ladley, R.P, Chatman, K, Mankin, A.S, Polikanov, Y.S, Myers, A.G. | | Deposit date: | 2021-08-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A synthetic antibiotic class overcoming bacterial multidrug resistance.
Nature, 599, 2021
|
|
8SKX
 
 | |
8SKE
 
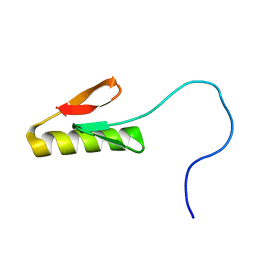 | |
8SKD
 
 | |
8SHM
 
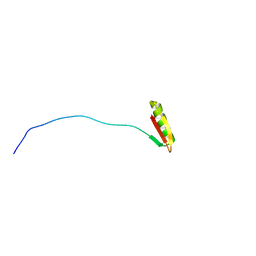 | |
7RQ9
 
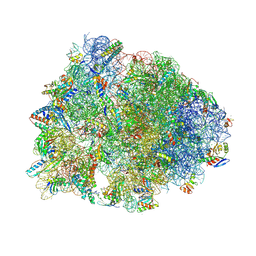 | | Crystal structure of the A2058-dimethylated Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAs, and aminoacylated P-site tRNA at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mitcheltree, M.J, Pisipati, A, Syroegin, E.A, Silvestre, K.J, Klepacki, D, Mason, J.D, Terwilliger, D.W, Testolin, G, Pote, A.R, Wu, K.J.Y, Ladley, R.P, Chatman, K, Mankin, A.S, Polikanov, Y.S, Myers, A.G. | | Deposit date: | 2021-08-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A synthetic antibiotic class overcoming bacterial multidrug resistance.
Nature, 599, 2021
|
|
4K7U
 
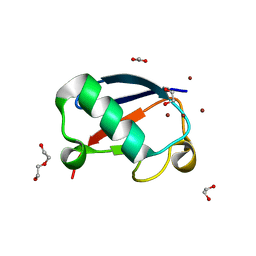 | | Crystal structure of Zn2.3-hUb (human ubiquitin) adduct from a solution 70 mM zinc acetate/1.3 mM hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fermani, S, Falini, G, Calvaresi, M, Bottoni, A, Arnesano, F, Natile, G. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Conformational selection of ubiquitin quaternary structures driven by zinc ions.
Chemistry, 19, 2013
|
|
2A55
 
 | | Solution structure of the two N-terminal CCP modules of C4b-binding protein (C4BP) alpha-chain. | | Descriptor: | C4b-binding protein | | Authors: | Jenkins, H.T, Mark, L, Ball, G, Lindahl, G, Uhrin, D, Blom, A.M, Barlow, P.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-12-13 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Human C4b-binding Protein, Structural Basis for Interaction with Streptococcal M Protein, a Major Bacterial Virulence Factor
J.Biol.Chem., 281, 2006
|
|
4K7W
 
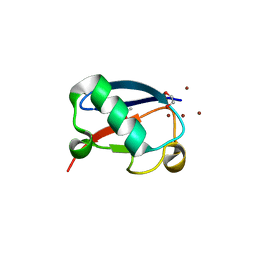 | | Crystal structure of Zn3-hUb(human ubiquitin) adduct from a solution 100 mM zinc acetate/1.3 mM hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ZINC ION, ... | | Authors: | Fermani, S, Falini, G, Calvaresi, M, Bottoni, A, Arnesano, F, Natile, G. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Conformational selection of ubiquitin quaternary structures driven by zinc ions.
Chemistry, 19, 2013
|
|
4K7S
 
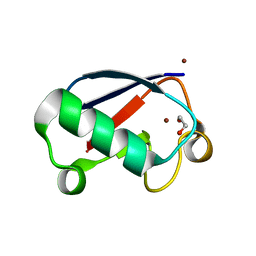 | | Crystal structure of Zn2-hUb (human ubiquitin) adduct from a solution 35 mM zinc acetate/1.3 mM hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ZINC ION, ... | | Authors: | Fermani, S, Falini, G, Calvaresi, M, Bottoni, A, Arnesano, F, Natile, G. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Conformational selection of ubiquitin quaternary structures driven by zinc ions.
Chemistry, 19, 2013
|
|
1RL0
 
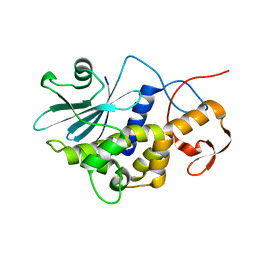 | | Crystal structure of a new ribosome-inactivating protein (RIP): dianthin 30 | | Descriptor: | Antiviral protein DAP-30 | | Authors: | Fermani, S, Falini, G, Ripamonti, A, Bolognesi, A, Polito, L, Stirpe, F. | | Deposit date: | 2003-11-24 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.4A structure of dianthin 30 indicates a role of surface potential at the active site of type 1 ribosome inactivating proteins
J.Struct.Biol., 149, 2005
|
|
6WL6
 
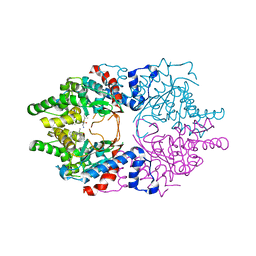 | | Cocomplex structure of Deoxyhypusine synthase with inhibitor 6-[(2R)-1-AMINO-4-METHYLPENTAN-2-YL]-3-(PYRIDIN-3-YL)-4H,5H,6H,7H-THIENO[2,3-C]PYRIDIN-7-ONE | | Descriptor: | 6-[(2R)-1-amino-4-methylpentan-2-yl]-3-(pyridin-3-yl)-5,6-dihydrothieno[2,3-c]pyridin-7(4H)-one, Deoxyhypusine synthase | | Authors: | Klein, M.G, Ambrus-Aikelin, G. | | Deposit date: | 2020-04-18 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | New Series of Potent Allosteric Inhibitors of Deoxyhypusine Synthase.
Acs Med.Chem.Lett., 11, 2020
|
|
6WKZ
 
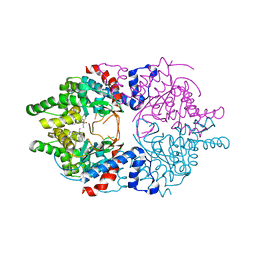 | | Cocomplex structure of Deoxyhypusine synthase with inhibitor 6-[(1R)-2-AMINO-1-PHENYLETHYL]-3-(PYRIDIN-3-YL)-4H,5H,6H,7H-THIENO[2,3-C]PYRIDIN-7-ONE | | Descriptor: | 6-[(1R)-2-amino-1-phenylethyl]-3-(pyridin-3-yl)-5,6-dihydrothieno[2,3-c]pyridin-7(4H)-one, Deoxyhypusine synthase | | Authors: | Klein, M.G, Ambrus-Aikelin, G. | | Deposit date: | 2020-04-17 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | New Series of Potent Allosteric Inhibitors of Deoxyhypusine Synthase.
Acs Med.Chem.Lett., 11, 2020
|
|
3K2B
 
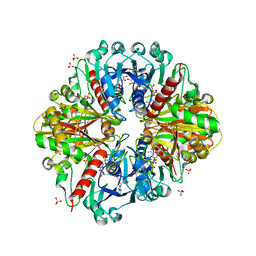 | | Crystal structure of photosynthetic A4 isoform glyceraldehyde-3-phosphate dehydrogenase complexed with NAD, from Arabidopsis thaliana | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Falini, G, Thumiger, A, Sparla, F, Marri, L, Trost, P. | | Deposit date: | 2009-09-29 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase (isoform A4) from Arabidopsis thaliana in complex with NAD
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7AAS
 
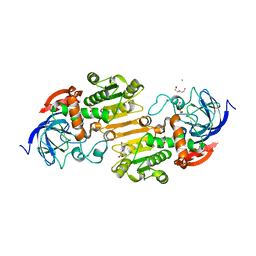 | | Crystal structure of nitrosoglutathione reductase (GSNOR) from Chlamydomonas reinhardtii | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, S-(hydroxymethyl)glutathione dehydrogenase, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Lemaire, S.D. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insights into nitrosoglutathione reductase from Chlamydomonas reinhardtii.
Redox Biol, 38, 2020
|
|
7AV7
 
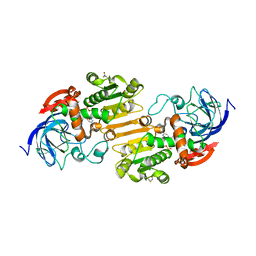 | | Crystal structure of S-nitrosylated nitrosoglutathione reductase(GSNOR)from Chlamydomonas reinhardtii, in complex with NAD+ | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-(hydroxymethyl)glutathione dehydrogenase, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Lemaire, S.D. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into nitrosoglutathione reductase from Chlamydomonas reinhardtii.
Redox Biol, 38, 2020
|
|
1R71
 
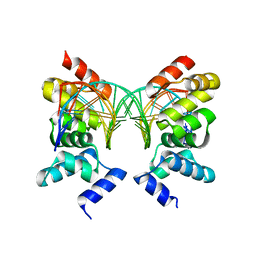 | | Crystal Structure of the DNA binding domain of KorB in complex with the operator DNA | | Descriptor: | 5'-D(*AP*(BRU)P*TP*TP*TP*AP*GP*CP*GP*GP*CP*TP*AP*AP*AP*AP*G)-3', 5'-D(*CP*(BRU)P*TP*TP*TP*AP*GP*CP*CP*GP*CP*TP*AP*AP*AP*AP*(BRU))-3', Transcriptional repressor protein korB | | Authors: | Khare, D, Ziegelin, G, Lanka, E, Heinemann, U. | | Deposit date: | 2003-10-17 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sequence-specific DNA binding determined by contacts outside the helix-turn-helix motif of the ParB homolog KorB.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SB1
 
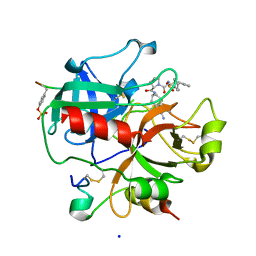 | | Novel Non-Covalent Thrombin Inhibitors Incorporating P1 4,5,6,7-Tetrahydrobenzothiazole Arginine Side Chain Mimetics | | Descriptor: | N-(BENZYLSULFONYL)-3-CYCLOHEXYLALANYL-N-(2-AMINO-1,3-BENZOTHIAZOL-6-YL)PROLINAMIDE, Prothrombin, SODIUM ION, ... | | Authors: | Marinko, P, Krbavcic, A, Mlinsek, G, Solmajer, T, Trampus-Bakija, A, Stegnar, M, Stojan, J, Kikelj, D. | | Deposit date: | 2004-02-09 | | Release date: | 2004-06-08 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel non-covalent thrombin inhibitors incorporating P(1) 4,5,6,7-tetrahydrobenzothiazole arginine side chain mimetics
Eur.J.Med.Chem., 39, 2004
|
|
1RG6
 
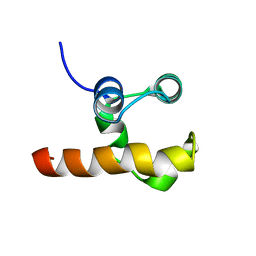 | | Solution structure of the C-terminal domain of p63 | | Descriptor: | second splice variant p63 | | Authors: | Cadot, B, Candi, E, Cicero, D.O, Desideri, A, Mele, S, Melino, G, Paci, M. | | Deposit date: | 2003-11-11 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of p63
To be Published
|
|
2PKR
 
 | | Crystal structure of (A+CTE)4 chimeric form of photosyntetic glyceraldehyde-3-phosphate dehydrogenase, complexed with NADP | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase Aor, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Fermani, S, Falini, G, Ripamonti, A. | | Deposit date: | 2007-04-18 | | Release date: | 2007-06-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism of thioredoxin regulation in photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3N30
 
 | | Crystal Structure of cubic Zn3-hUb (human ubiquitin) adduct | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Siliqi, D, Caliandro, R, Arnesano, F, Natile, G, Falini, G, Fermani, S, Belviso, B.D. | | Deposit date: | 2010-05-19 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analysis of metal-ion binding to human ubiquitin.
Chemistry, 17, 2011
|
|
