3C3G
 
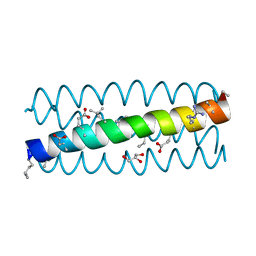 | | alpha/beta-Peptide helix bundles: The GCN4-pLI side chain sequence on an (alpha-alpha-beta) backbone | | Descriptor: | GLYCEROL, alpha/beta peptide with the GCN4-pLI side chain sequence on an (alpha-alpha-beta) backbone | | Authors: | Horne, W.S, Price, J.L, Gellman, S.H. | | Deposit date: | 2008-01-28 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interplay among side chain sequence, backbone composition, and residue rigidification in polypeptide folding and assembly.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BOF
 
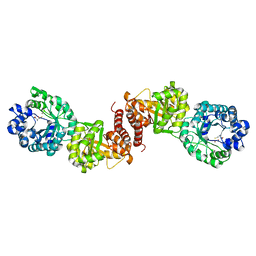 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima complexed with Zn2+ and Homocysteine | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydrofolate S-homocysteine methyltransferase, POTASSIUM ION, ... | | Authors: | Koutmos, M, Smith, J.L, Ludwig, M.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal active site elasticity linked to activation of homocysteine in methionine synthases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CAU
 
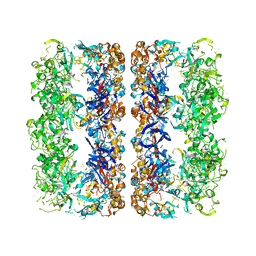 | | D7 symmetrized structure of unliganded GroEL at 4.2 Angstrom resolution by cryoEM | | Descriptor: | 60 kDa chaperonin | | Authors: | Ludtke, S.J, Baker, M.L, Chen, D.H, Song, J.L, Chuang, D, Chiu, W. | | Deposit date: | 2008-02-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | De Novo Backbone Trace of GroEL from Single Particle Electron Cryomicroscopy.
Structure, 16, 2008
|
|
3C94
 
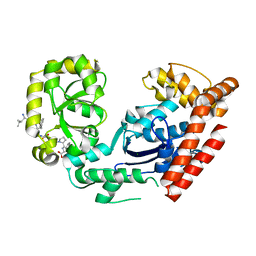 | | ExoI/SSB-Ct complex | | Descriptor: | Exodeoxyribonuclease I, MAGNESIUM ION, Single-stranded DNA-binding C-terminal tail peptide | | Authors: | Lu, D, Keck, J.L. | | Deposit date: | 2008-02-15 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of Escherichia coli single-stranded DNA-binding protein stimulation of exonuclease I.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3CE6
 
 | | Crystal structure of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with NAD and adenosine | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Reddy, M.C.M, Gokulan, K, Shetty, N.D, Owen, J.L, Ioerger, T.R, Sacchettini, J.C. | | Deposit date: | 2008-02-28 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with substrate and inhibitors.
Protein Sci., 17, 2008
|
|
3C9B
 
 | | Crystal structure of SeMet Vps75 | | Descriptor: | Vacuolar protein sorting-associated protein 75 | | Authors: | Keck, J.L, Berndsen, C.E, Tsubota, T, Lindner, S.E, Lee, S, Holton, J.M, Kaufman, P.D, Denu, J.M. | | Deposit date: | 2008-02-15 | | Release date: | 2008-08-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75
Nat.Struct.Mol.Biol., 15, 2008
|
|
3C9V
 
 | | C7 Symmetrized Structure of Unliganded GroEL at 4.7 Angstrom Resolution from CryoEM | | Descriptor: | 60 kDa chaperonin | | Authors: | Ludtke, S.J, Baker, M.L, Chen, D.H, Song, J.L, Chuang, D, Chiu, W. | | Deposit date: | 2008-02-18 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | De Novo Backbone Trace of GroEL from Single Particle Electron Cryomicroscopy.
Structure, 16, 2008
|
|
6ECW
 
 | | StiD O-MT residues 956-1266 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, StiD protein | | Authors: | Skiba, M.A, Bivins, M.M, Smith, J.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Polyketide Synthase O-Methylation.
ACS Chem. Biol., 13, 2018
|
|
6EJJ
 
 | | Structure of a glycosyltransferase / state 2 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, NerylNeryl pyrophosphate, ... | | Authors: | Ramirez, A.S, Boilevin, J, Mehdipour, A.R, Hummer, G, Darbre, T, Reymond, J.L, Locher, K.P. | | Deposit date: | 2017-09-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the molecular ruler mechanism of a bacterial glycosyltransferase.
Nat Commun, 9, 2018
|
|
6E3K
 
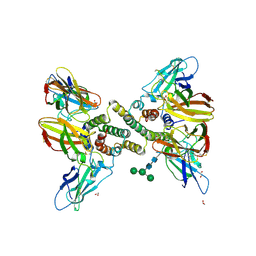 | | Interferon gamma signalling complex with IFNGR1 and IFNGR2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jude, K.M, Mendoza, J.L, Garcia, K.C. | | Deposit date: | 2018-07-14 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the IFN gamma receptor complex guides design of biased agonists.
Nature, 567, 2019
|
|
6E3L
 
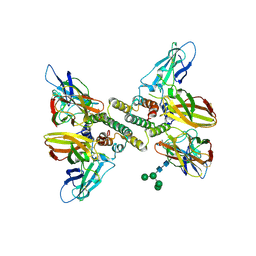 | | Interferon gamma signalling complex with IFNGR1 and IFNGR2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CYSTEINE, ... | | Authors: | Jude, K.M, Mendoza, J.L, Garcia, K.C. | | Deposit date: | 2018-07-14 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the IFN gamma receptor complex guides design of biased agonists.
Nature, 567, 2019
|
|
6ECX
 
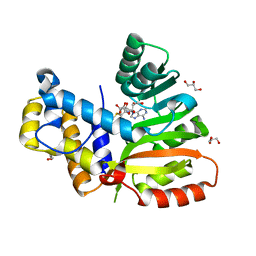 | | StiE O-MT residues 942-1257 | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, StiE protein | | Authors: | Skiba, M.A, Bivins, M.B, Smith, J.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Polyketide Synthase O-Methylation.
ACS Chem. Biol., 13, 2018
|
|
5YDP
 
 | |
5ZJL
 
 | |
5ZXH
 
 | | The structure of MT189-tubulin complex | | Descriptor: | 2-(6-fluoro-3-{[(4-methoxyphenyl)methyl]amino}imidazo[1,2-a]pyridin-2-yl)phenol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Li, Z.P, Wang, Y.X, Meng, T, Yang, J.L, Chen, Q. | | Deposit date: | 2018-05-21 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of MT189-Tubulin Complex Provides Insights into Drug Design
Lett.Drug Des.Discovery, 2019
|
|
6AL7
 
 | | Crystal structure HpiC1 F138S | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.687 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
7P9U
 
 | | Cryo EM structure of System XC- in complex with glutamate | | Descriptor: | 4F2 cell-surface antigen heavy chain, Cystine/glutamate transporter, GLUTAMIC ACID | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-17 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for redox control by the human cystine/glutamate antiporter system xc .
Nat Commun, 12, 2021
|
|
7P9V
 
 | | Cryo EM structure of System XC- | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, Cystine/glutamate transporter | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-17 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for redox control by the human cystine/glutamate antiporter system xc .
Nat Commun, 12, 2021
|
|
7OXE
 
 | |
4UBE
 
 | | CRYSTAL STRUCTURE OF M TUBERCULOSIS ADENOSINE KINASE COMPLEXED WITH 2-FLURO ADENOSINE | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Adenosine kinase | | Authors: | Reddy, M.C.M, Palaninathan, S.K, Shetty, N.D, Owen, J.L, Watson, M.D, Sacchettini, J.C. | | Deposit date: | 2014-08-12 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | CRYSTAL STRUCTURE OF M TUBERCULOSIS ADENOSINE KINASE COMPLEXED WITH 2-FLURO ADENOSINE
TO BE PUBLISHED
|
|
7Q21
 
 | | III2-IV2 respiratory supercomplex from Corynebacterium glutamicum | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Kovalova, T, Moe, A, Krol, S, Yanofsky, D.J, Bott, M, Sjostrand, D, Rubinstein, J.L, Hogbom, M, Brzezinski, P. | | Deposit date: | 2021-10-22 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The respiratory supercomplex from C. glutamicum.
Structure, 30, 2022
|
|
1MHM
 
 | |
1MMI
 
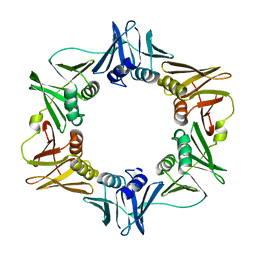 | | E. COLI DNA POLYMERASE BETA SUBUNIT | | Descriptor: | DNA polymerase III, beta chain | | Authors: | Oakley, A.J, Prosselkov, P, Wijffels, G, Beck, J.L, Wilce, M.C.J, Dixon, N.E. | | Deposit date: | 2002-09-04 | | Release date: | 2003-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Flexibility revealed by the 1.85 A crystal structure of the beta sliding-clamp subunit of Escherichia coli DNA polymerase III.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6XSG
 
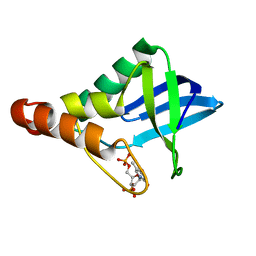 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66T at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Robinson, A.C, Schlessman, J.L, Garcia-Moreno E, B, Sorenson, J.L. | | Deposit date: | 2020-07-15 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66T at cryogenic temperature
To be Published
|
|
1MPJ
 
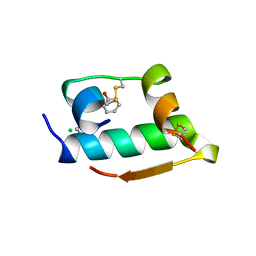 | | X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL | | Descriptor: | CHLORIDE ION, PHENOL, PHENOL INSULIN, ... | | Authors: | Whittingham, J.L, Dodson, E.J, Moody, P.C.E, Dodson, G.G. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystallographic studies on hexameric insulins in the presence of helix-stabilizing agents, thiocyanate, methylparaben, and phenol.
Biochemistry, 34, 1995
|
|
