6XJH
 
 | | PmtCD ABC exporter without the basket domain at C2 symmetry | | Descriptor: | ABC transporter ATP-binding protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zeytuni, N, Strynadka, N.J.C, Hu, J, Worrall, L.J, Chou, H, Yu, Z. | | Deposit date: | 2020-06-23 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insight into the Staphylococcus aureus ATP-driven exporter of virulent peptide toxins
Sci Adv, 6, 2020
|
|
1XZ0
 
 | | Crystal structure of CD1a in complex with a synthetic mycobactin lipopeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(HYDROXY-HEXADECANOYL-AMINO)-2-{[(4S)-2-(2-HYDROXY-PHENYL)-4,5-DIHYDRO-OXAZOLE-4-CARBONYL]-AMINO}-HEXANOIC ACID 2-[(3S)-1-(TERT-BUTYL-DIPHENYL-SILANYLOXY)-2-OXO-AZEPAN-3-YLCARBAMOYL]-(1S)-1-METHYL-ETHYL ESTER, Beta-2-microglobulin, ... | | Authors: | Zajonc, D.M, Crispin, M.D, Bowden, T.A, Young, D.C, Cheng, T.Y, Hu, J, Costello, C.E, Miller, M.J, Moody, D.B, Wilson, I.A. | | Deposit date: | 2004-11-11 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Mechanism of Lipopeptide Presentation by CD1a.
Immunity, 22, 2005
|
|
2AUH
 
 | | Crystal structure of the Grb14 BPS region in complex with the insulin receptor tyrosine kinase | | Descriptor: | CALCIUM ION, Growth factor receptor-bound protein 14, Insulin receptor | | Authors: | Depetris, R.S, Hu, J, Gimpelevich, I, Holt, L.J, Daly, R.J, Hubbard, S.R. | | Deposit date: | 2005-08-27 | | Release date: | 2005-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for inhibition of the insulin receptor by the adaptor protein grb14.
Mol.Cell, 20, 2005
|
|
2AUG
 
 | | Crystal structure of the Grb14 SH2 domain | | Descriptor: | Growth factor receptor-bound protein 14 | | Authors: | Depetris, R.S, Hu, J, Gimpelevich, I, Holt, L.J, Daly, R.J, Hubbard, S.R. | | Deposit date: | 2005-08-27 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for inhibition of the insulin receptor by the adaptor protein grb14.
Mol.Cell, 20, 2005
|
|
5C7M
 
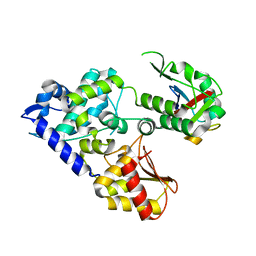 | | CRYSTAL STRUCTURE OF E3 LIGASE ITCH WITH A UB VARIANT | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Polyubiquitin-C | | Authors: | Walker, J.R, Hu, J, Dong, A, Wernimont, A, Zhang, W, Sidhu, S, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
5C7J
 
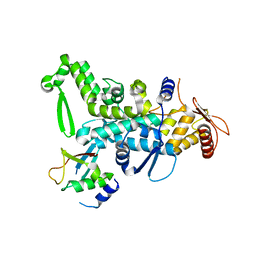 | | CRYSTAL STRUCTURE OF NEDD4 WITH A UB VARIANT | | Descriptor: | E3 ubiquitin-protein ligase NEDD4, Polyubiquitin-C | | Authors: | Walker, J.R, Hu, J, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
3FP2
 
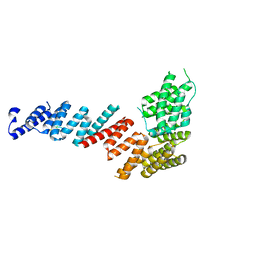 | | Crystal structure of Tom71 complexed with Hsp82 C-terminal fragment | | Descriptor: | ATP-dependent molecular chaperone HSP82, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Li, J, Qian, X, Hu, J, Sha, B. | | Deposit date: | 2009-01-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading.
J.Biol.Chem., 284, 2009
|
|
3FP3
 
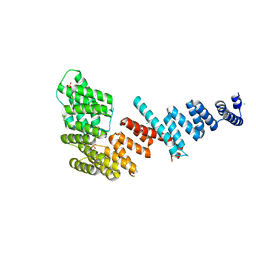 | | Crystal structure of Tom71 | | Descriptor: | CHLORIDE ION, SULFATE ION, TPR repeat-containing protein YHR117W | | Authors: | Li, J, Qian, X, Hu, J, Sha, B. | | Deposit date: | 2009-01-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading.
J.Biol.Chem., 284, 2009
|
|
3FP4
 
 | | Crystal structure of Tom71 complexed with Ssa1 C-terminal fragment | | Descriptor: | CHLORIDE ION, SODIUM ION, SULFATE ION, ... | | Authors: | Li, J, Qian, X, Hu, J, Sha, B. | | Deposit date: | 2009-01-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading.
J.Biol.Chem., 284, 2009
|
|
6UTR
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase in complex with copper | | Descriptor: | ATP-dependent sacrificial sulfur transferase LarE, COPPER (II) ION, PHOSPHATE ION, ... | | Authors: | Fellner, M, Huizenga, K, Hausinger, R.P, Hu, J. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystallographic characterization of a tri-Asp metal-binding site at the three-fold symmetry axis of LarE.
Sci Rep, 10, 2020
|
|
8UC2
 
 | | Ethylene forming enzyme (EFE) R171A variant in complex with nickel and Benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, BENZOIC ACID, ... | | Authors: | Chatterjee, S, Rankin, J.A, Hu, J, Hausinger, R. | | Deposit date: | 2023-09-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, Spectroscopic, and Computational Insights from Canavanine-Bound and Two Catalytically Compromised Variants of the Ethylene-Forming Enzyme.
Biochemistry, 63, 2024
|
|
6UTQ
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase in complex with cadmium | | Descriptor: | ATP-dependent sacrificial sulfur transferase LarE, CADMIUM ION, PHOSPHATE ION, ... | | Authors: | Fellner, M, Huizenga, K, Hausinger, R.P, Hu, J. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystallographic characterization of a tri-Asp metal-binding site at the three-fold symmetry axis of LarE.
Sci Rep, 10, 2020
|
|
6UTT
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase in complex with calcium | | Descriptor: | ATP-dependent sacrificial sulfur transferase LarE, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Fellner, M, Huizenga, K, Hausinger, R.P, Hu, J. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystallographic characterization of a tri-Asp metal-binding site at the three-fold symmetry axis of LarE.
Sci Rep, 10, 2020
|
|
4RXX
 
 | | Crystal Structure of the N-terminal Domain of Human Ubiquitin Specific Protease 38 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Shen, L, Hu, J, Li, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-12 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the N-terminal Domain of Human Ubiquitin Specific Protease 38
to be published
|
|
6UTP
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase in complex with cobalt | | Descriptor: | ATP-dependent sacrificial sulfur transferase LarE, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Fellner, M, Huizenga, K, Hausinger, R.P, Hu, J. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Crystallographic characterization of a tri-Asp metal-binding site at the three-fold symmetry axis of LarE.
Sci Rep, 10, 2020
|
|
8SOQ
 
 | | S127A variant of LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with authentic substrate NaAD | | Descriptor: | MAGNESIUM ION, NICOTINIC ACID ADENINE DINUCLEOTIDE, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase | | Authors: | Chatterjee, S, Rankin, J.A, Hu, J, Hausinger, R.P. | | Deposit date: | 2023-04-29 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the LarB-Substrate Complex and Identification of a Reaction Intermediate during Nickel-Pincer Nucleotide Cofactor Biosynthesis.
Biochemistry, 62, 2023
|
|
8STD
 
 | | S127A variant of LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with authentic substrate NaAD and soaked with CS2 | | Descriptor: | MAGNESIUM ION, NICOTINIC ACID ADENINE DINUCLEOTIDE, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase | | Authors: | Chatterjee, S, Rankin, J.A, Hu, J, Hausinger, R.P. | | Deposit date: | 2023-05-10 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the LarB-Substrate Complex and Identification of a Reaction Intermediate during Nickel-Pincer Nucleotide Cofactor Biosynthesis.
Biochemistry, 62, 2023
|
|
8STC
 
 | | S127A variant of LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with Zinc and soaked with bicarbonate. | | Descriptor: | MAGNESIUM ION, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase, ZINC ION | | Authors: | Chatterjee, S, Rankin, J.A, Hu, J, Hausinger, R.P. | | Deposit date: | 2023-05-09 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | S127A variant of LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with Zinc and soaked with bicarbonate.
To be published
|
|
5FBK
 
 | | Crystal structure of the extracellular domain of human calcium sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Zhang, T, Zhang, C, Miller, C.L, Zou, J, Moremen, K.W, Brown, E.M, Yang, J.J, Hu, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist.
Sci Adv, 2, 2016
|
|
5FBH
 
 | | Crystal structure of the extracellular domain of human calcium sensing receptor with bound Gd3+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Zhang, T, Zhang, C, Miller, C.L, Zou, J, Moremen, K.W, Brown, E.M, Yang, J.J, Hu, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist.
Sci Adv, 2, 2016
|
|
3NYJ
 
 | | Crystal Structure Analysis of APP E2 domain | | Descriptor: | Amyloid beta A4 protein, OSMIUM ION | | Authors: | Ha, Y, Hu, J, Lee, S, Liu, X. | | Deposit date: | 2010-07-15 | | Release date: | 2011-06-01 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The E2 Domains of APP and APLP1 Share a Conserved Mode of Dimerization.
Biochemistry, 50, 2011
|
|
7WNT
 
 | | RNase J from Mycobacterium tuberculosis | | Descriptor: | Ribonuclease J, ZINC ION | | Authors: | Li, J, Bao, L, Hu, J, Zhan, B, Li, Z. | | Deposit date: | 2022-01-19 | | Release date: | 2023-01-25 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural insights into RNase J that plays an essential role in Mycobacterium tuberculosis RNA metabolism
Nat Commun, 14, 2023
|
|
7WNU
 
 | | Mycobacterium tuberculosis Rnase J complex with 7nt RNA | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*AP*AP*A)-3'), Ribonuclease J, ZINC ION | | Authors: | Li, J, Hu, J, Bao, L, Zhan, B, Li, Z. | | Deposit date: | 2022-01-19 | | Release date: | 2023-01-25 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into RNase J that plays an essential role in Mycobacterium tuberculosis RNA metabolism
Nat Commun, 14, 2023
|
|
4DAJ
 
 | | Structure of the M3 Muscarinic Acetylcholine Receptor | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, Muscarinic acetylcholine receptor M3, Lysozyme, ... | | Authors: | Kruse, A.C, Hu, J, Pan, A.C, Arlow, D.H, Rosenbaum, D.M, Rosemond, E, Green, H.F, Liu, T, Chae, P.S, Dror, R.O, Shaw, D.E, Weis, W.I, Wess, J, Kobilka, B. | | Deposit date: | 2012-01-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and dynamics of the M3 muscarinic acetylcholine receptor.
Nature, 482, 2012
|
|
6CF3
 
 | | Ethylene forming enzyme Y306A variant in complex with manganese and 2-oxoglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ... | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2018-02-13 | | Release date: | 2019-02-13 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural, Spectroscopic, and Computational Insights from Canavanine-Bound and Two Catalytically Compromised Variants of the Ethylene-Forming Enzyme.
Biochemistry, 2024
|
|
