5ZQI
 
 | | Alginate lyase AlgAT5 from Polysaccharide Lyase family 7 | | Descriptor: | Alginate lyase AlgAT5, SULFATE ION | | Authors: | Su, H, Dong, S, Feng, Y.G, Ji, S.Q, Lu, M, Li, F.L. | | Deposit date: | 2018-04-19 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Alginate lyase AlgAT5 from Polysaccharide Lyase family 7
To Be Published
|
|
7EGN
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | H-Bonding Networks Dictate the Molecular Mechanism of H2O2 Activation by P450
Acs Catalysis, 11, 2021
|
|
8HDJ
 
 | |
8HOT
 
 | |
8HOO
 
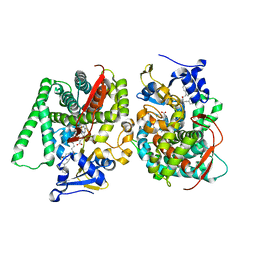 | |
7WDH
 
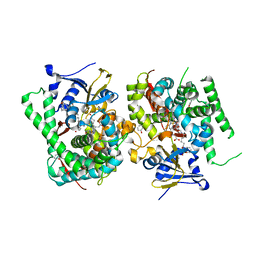 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine, phenol and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Engineering Cytochrome P450BM3 Enzymes for Direct Nitration of Unsaturated Hydrocarbons.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7WDG
 
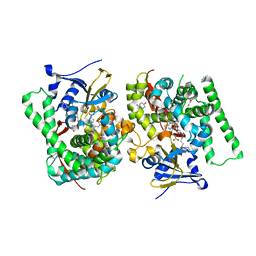 | | Crystal structure of the P450 BM3 heme domain mutant F87L in complex with N-imidazolyl-hexanoyl-L-phenylalanine, phenol and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Engineering Cytochrome P450BM3 Enzymes for Direct Nitration of Unsaturated Hydrocarbons.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
1KHY
 
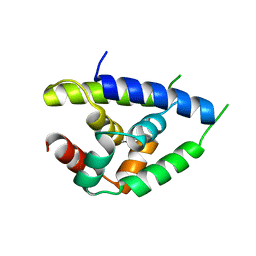 | |
6LUS
 
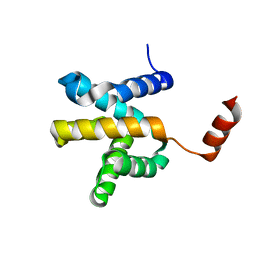 | | Crystal structure of the Mengla Virus VP30 C-terminal domain | | Descriptor: | Minor nucleoprotein VP30 | | Authors: | Dong, S.S, Wen, K.N, Chu, H.G, Wang, C.H, Qin, X.C. | | Deposit date: | 2020-01-30 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the Mengla virus VP30 C-terminal domain.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
4Z9P
 
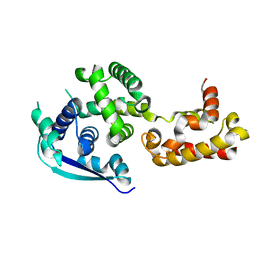 | | Crystal structure of Ebola virus nucleoprotein core domain at 1.8A resolution | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Dong, S.S, Yang, P, Li, G.B, Liu, B.C, Yang, C, Rao, Z.H. | | Deposit date: | 2015-04-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Insight into the Ebola virus nucleocapsid assembly mechanism: crystal structure of Ebola virus nucleoprotein core domain at 1.8 A resolution.
Protein Cell, 6, 2015
|
|
7VZG
 
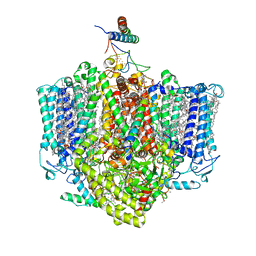 | | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c (the larger form) | | Descriptor: | BACTERIOCHLOROPHYLL A, CALCIUM ION, CHLOROPHYLL A, ... | | Authors: | Huang, G.Q, Dong, S.S, Qin, X.C, Sui, S.F. | | Deposit date: | 2021-11-16 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c.
Nat Commun, 13, 2022
|
|
7VZR
 
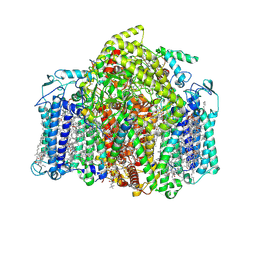 | | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c (the smaller form) | | Descriptor: | BACTERIOCHLOROPHYLL A, CALCIUM ION, CHLOROPHYLL A, ... | | Authors: | Huang, G.Q, Dong, S.S, Qin, X.C, Sui, S.F. | | Deposit date: | 2021-11-16 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.22 Å) | | Cite: | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c
Nat Commun, 13, 2022
|
|
7UYA
 
 | | Inhibitor bound VIM1 | | Descriptor: | (2M)-4'-(piperidin-4-yl)-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYD
 
 | | Inhibitor bound VIM1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYB
 
 | | Inhibitor bound VIM1 | | Descriptor: | (2M)-4'-(piperidin-4-yl)-2-(1H-tetrazol-5-yl)-4-(trifluoromethyl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYC
 
 | | Inhibitor bound VIM1 | | Descriptor: | (2P)-4'-(piperidin-4-yl)-4-[(piperidin-4-yl)methyl]-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, MAGNESIUM ION, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
6RYB
 
 | |
6RYA
 
 | | Structure of Dup1 mutant H67A:Ubiquitin complex | | Descriptor: | Polyubiquitin-C, Septation initiation protein | | Authors: | Donghyuk, S, Ivan, D. | | Deposit date: | 2019-06-10 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Regulation of Phosphoribosyl-Linked Serine Ubiquitination by Deubiquitinases DupA and DupB.
Mol.Cell, 77, 2020
|
|
9C5P
 
 | | Inhibitor bound VIM1 | | Descriptor: | (2M)-4-(piperidine-4-sulfonyl)-4'-(piperidin-4-yl)-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, MAGNESIUM ION, Metallo-beta-lactamase type 2, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2024-06-06 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Discovery of sulfone containing metallo-beta-lactamase inhibitors with reduced bacterial cell efflux and histamine release issues.
Bioorg.Med.Chem.Lett., 114, 2024
|
|
6MPZ
 
 | | Crystal structure of a double glycine motif protease from AMS/PCAT transporter in complex with the leader peptide | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE, Double Glycine Motif Protease domain from AMS/PCAT Transporter, peptide aldehyde inhibitor 1 based on the ProcA2.8 leader peptide | | Authors: | Dong, S.-H, Nair, S.K. | | Deposit date: | 2018-10-09 | | Release date: | 2019-02-06 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into AMS/PCAT transporters from biochemical and structural characterization of a double Glycine motif protease.
Elife, 8, 2019
|
|
6M6B
 
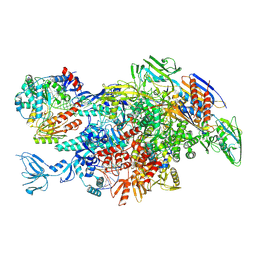 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase and ATP-gamma-S | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6M6A
 
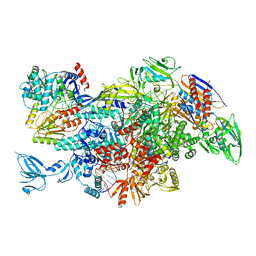 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6CIB
 
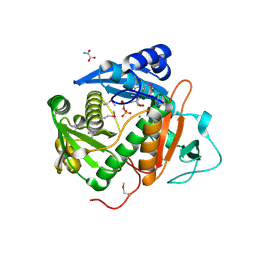 | |
6CI7
 
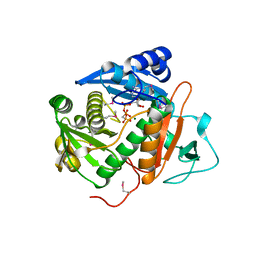 | |
6YHL
 
 | |
