2TLD
 
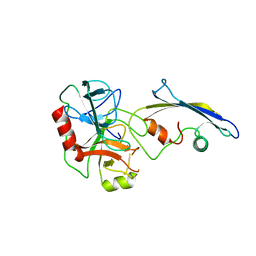 | | CRYSTAL STRUCTURE OF AN ENGINEERED SUBTILISIN INHIBITOR COMPLEXED WITH BOVINE TRYPSIN | | Descriptor: | STREPTOMYCES SUBTILISIN INHIBITOR (SSI), TRYPSIN | | Authors: | Mitsui, Y, Takeuchi, Y, Nonaka, T, Nakamura, K.T. | | Deposit date: | 1991-09-16 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an engineered subtilisin inhibitor complexed with bovine trypsin.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
2RDV
 
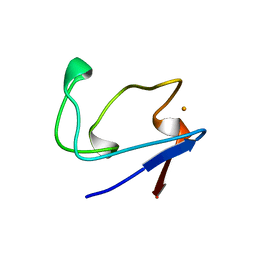 | |
7YRU
 
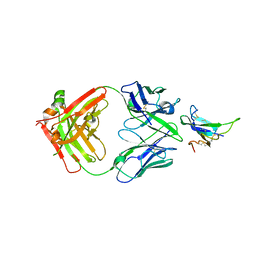 | | ALK2 antibody complex | | Descriptor: | Activin receptor type-1, antibody heavy chain, antibody light chain | | Authors: | Kawaguchi, Y, Nakamura, K, Suzuki, M, Tsuji, S, Katagiri, T. | | Deposit date: | 2022-08-10 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A blocking monoclonal antibody reveals dimerization of intracellular domains of ALK2 associated with genetic disorders.
Nat Commun, 14, 2023
|
|
2SIC
 
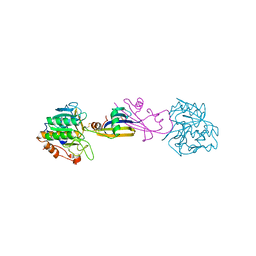 | | REFINED CRYSTAL STRUCTURE OF THE COMPLEX OF SUBTILISIN BPN' AND STREPTOMYCES SUBTILISIN INHIBITOR AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, STREPTOMYCES SUBTILISIN INHIBITOR (SSI), SUBTILISIN BPN' | | Authors: | Mitsui, Y, Takeuchi, Y, Hirono, S, Akagawa, H, Nakamura, K.T. | | Deposit date: | 1991-04-01 | | Release date: | 1993-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined crystal structure of the complex of subtilisin BPN' and Streptomyces subtilisin inhibitor at 1.8 A resolution.
J.Mol.Biol., 221, 1991
|
|
3SIC
 
 | |
7VSX
 
 | | Crystal structure of QL-nanoKAZ (Reverse mutant of nanoKAZ with L18Q and V27L) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, QLnK | | Authors: | Tomabechi, Y, Sekine, S, Shirouzu, M, Takamitsu, H, Satoshi, I. | | Deposit date: | 2021-10-27 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Reverse mutants of the catalytic 19 kDa mutant protein (nanoKAZ/nanoLuc) from Oplophorus luciferase with coelenterazine as preferred substrate.
Plos One, 17, 2022
|
|
7DHL
 
 | | Crystal structure of FGFR3 in complex with pyrimidine derivative | | Descriptor: | 5-[2-(3,5-dimethoxyphenyl)ethyl]-N-[3-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]pyrimidin-2-amine, Fibroblast growth factor receptor 3 | | Authors: | Echizen, Y, Tateishi, Y, Amano, Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Synthesis and structure-activity relationships of pyrimidine derivatives as potent and orally active FGFR3 inhibitors with both increased systemic exposure and enhanced in vitro potency.
Bioorg.Med.Chem., 33, 2021
|
|
5ZJ6
 
 | | Crystal structure of HCK kinase complexed with a pyrrolo-pyrimidine inhibitor 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2018-03-19 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Phosphorylated and non-phosphorylated HCK kinase domains produced by cell-free protein expression.
Protein Expr. Purif., 150, 2018
|
|
1RDV
 
 | |
5YHS
 
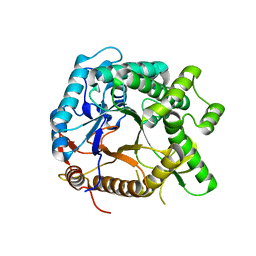 | | Pyruvylated beta-D-galactosidase from Bacillus sp. HMA207, apo form | | Descriptor: | Pyruvylated beta-D-galactosidase | | Authors: | Tanuma, M, Yamada, C, Arakawa, T, Higuchi, Y, Takegawa, K, Fushinobu, S. | | Deposit date: | 2017-09-29 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification and characterization of a novel beta-D-galactosidase that releases pyruvylated galactose.
Sci Rep, 8, 2018
|
|
5YIF
 
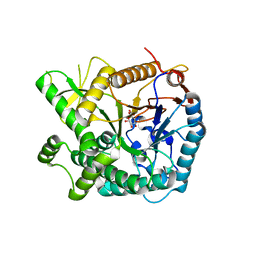 | | Pyruvylated beta-D-galactosidase from Bacillus sp. HMA207, E163A mutant pyruvylated beta-D-galactose complex | | Descriptor: | (2R,4aR,6R,7R,8R,8aR)-2-methyl-6,7,8-tris(oxidanyl)-4,4a,6,7,8,8a-hexahydropyrano[3,2-d][1,3]dioxine-2-carboxylic acid, Pyruvylated beta-D-galactosidase | | Authors: | Tanuma, M, Yamada, C, Arakawa, T, Higuchi, Y, Takegawa, K, Fushinobu, S. | | Deposit date: | 2017-10-04 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification and characterization of a novel beta-D-galactosidase that releases pyruvylated galactose.
Sci Rep, 8, 2018
|
|
2EWK
 
 | |
7C03
 
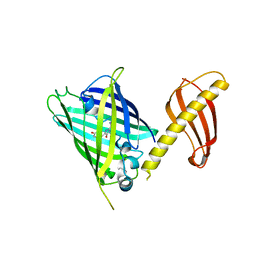 | | Crystal structure of POLArISact(T57S), genetically encoded probe for fluorescent polarization | | Descriptor: | POLArISact(T57S) | | Authors: | Tomabechi, Y, Sakai, N, Shirouzu, M. | | Deposit date: | 2020-04-30 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | POLArIS, a versatile probe for molecular orientation, revealed actin filaments associated with microtubule asters in early embryos.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2FFN
 
 | |
7EG2
 
 | | Crystal structure of the apoAequorin complex with (S)-daCTZ | | Descriptor: | (2~{S})-2-(hydroxymethyl)-6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-4-(phenylmethyl)-3~{H}-inden-1-one, Aequorin-2 | | Authors: | Tomabechi, Y, Shirouzu, M. | | Deposit date: | 2021-03-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Chiral deaza-coelenterazine analogs for probing a substrate-binding site in the Ca2+-binding photoprotein aequorin.
Plos One, 16, 2021
|
|
7EG3
 
 | | Crystal structure of the apoAequorin complex with (S)-HM-daCTZ | | Descriptor: | (2~{S})-6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-4-(phenylmethyl)-2,3-dihydroinden-1-one, Aequorin-2 | | Authors: | Tomabechi, Y, Shirouzu, M. | | Deposit date: | 2021-03-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Chiral deaza-coelenterazine analogs for probing a substrate-binding site in the Ca2+-binding photoprotein aequorin.
Plos One, 16, 2021
|
|
6IGU
 
 | | Crystal structure of the hydrolytic antibody Fab 9C10 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, IMMUNOGLOBULIN 9C10 H CHAIN, IMMUNOGLOBULIN 9C10 L CHAIN, ... | | Authors: | Yamaguchi, A, Tada, T, Tsuchiya, Y, Tsumuraya, T, Fujii, I. | | Deposit date: | 2018-09-26 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of the complex of the hydrolytic antibody Fab 9C10 and a transition-state analog
To Be Published
|
|
2FMY
 
 | |
5H09
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-ethyl2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanoate | | Descriptor: | Tyrosine-protein kinase HCK, ethyl (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-4-methyl-pentanoate | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0B
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanoic acid | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]azaniumyl]-4-methyl-pentanoate, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0H
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-N,N,4-trimethylpentanamide | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-~{N},~{N},4-trimethyl-pentanamide, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0E
 
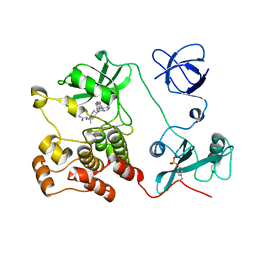 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanamide | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-4-methyl-pentanamide, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0G
 
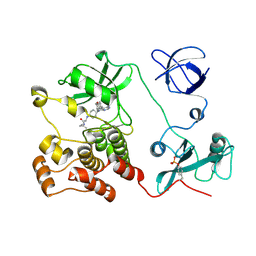 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-N,4-dimethylpentanamide | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-~{N},4-dimethyl-pentanamide, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5GV0
 
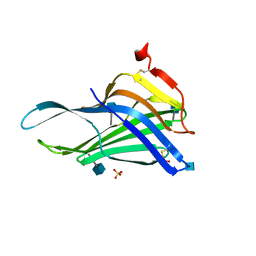 | | Crystal structure of the membrane-proximal domain of mouse lysosome-associated membrane protein 1 (LAMP-1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome-associated membrane glycoprotein 1, SULFATE ION | | Authors: | Tomabechi, Y, Ehara, H, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-09-01 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Lysosome-associated membrane proteins-1 and -2 (LAMP-1 and LAMP-2) assemble via distinct modes
Biochem.Biophys.Res.Commun., 479, 2016
|
|
5GV3
 
 | | Crystal structure of the membrane-distal domain of mouse lysosome-associated membrane protein 2 (LAMP-2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome-associated membrane glycoprotein 2, ZINC ION | | Authors: | Tomabechi, Y, Ehara, H, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-09-01 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Lysosome-associated membrane proteins-1 and -2 (LAMP-1 and LAMP-2) assemble via distinct modes.
Biochem. Biophys. Res. Commun., 479, 2016
|
|
