2OWI
 
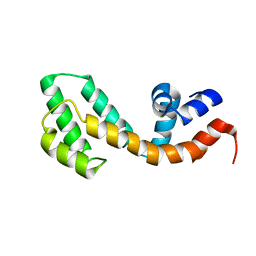 | | Solution structure of the RGS domain from human RGS18 | | Descriptor: | Regulator of G-protein signaling 18 | | Authors: | Higman, V.A, Leidert, M, Bray, J, Elkins, J, Soundararajan, M, Doyle, D.A, Gileadi, C, Phillips, C, Schoch, G, Yang, X, Brockmann, C, Schmieder, P, Diehl, A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Oschkinat, H, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-16 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2JAV
 
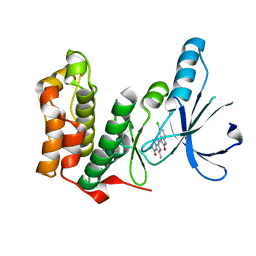 | | Human Kinase with pyrrole-indolinone ligand | | Descriptor: | 5-[(Z)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Pike, A.C.W, Rellos, P, Das, S, Fedorov, O, Papagrigoriou, E, Debreczeni, J.E, Turnbull, A.P, Gorrec, F, Bray, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A, Weigelt, J, von Delft, F, Knapp, S. | | Deposit date: | 2006-11-30 | | Release date: | 2006-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Regulation of the Human Nek2 Centrosomal Kinase
J.Biol.Chem., 282, 2007
|
|
2GZV
 
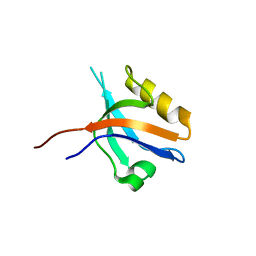 | | The cystal structure of the PDZ domain of human PICK1 | | Descriptor: | PRKCA-binding protein | | Authors: | Debreczeni, J.E, Elkins, J.M, Yang, X, Berridge, G, Bray, J, Colebrook, S, Smee, C, Savitsky, P, Gileadi, O, Turnbull, A, von Delft, F, Doyle, D.A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-12 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
2FCF
 
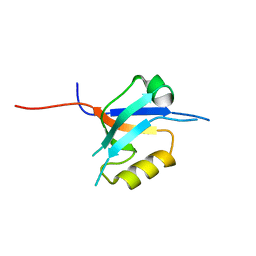 | | The crystal structure of the 7th PDZ domain of MPDZ (MUPP-1) | | Descriptor: | Multiple PDZ domain protein | | Authors: | Papagrigoriou, E, Berridge, G, Johansson, C, Colebrook, S, Salah, E, Burgess, N, Smee, C, Savitsky, P, Bray, J, Schoch, G, Phillips, C, Gileadi, C, Soundarajan, M, Yang, X, Elkins, J.M, Gorrec, F, Turnbull, A, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
2FNE
 
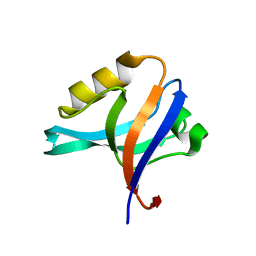 | | The crystal structure of the 13th PDZ domain of MPDZ | | Descriptor: | Multiple PDZ domain protein | | Authors: | Papagrigoriou, E, Berridge, G, Johansson, C, Colebrook, S, Salah, E, Burgess, N, Smee, C, Savitsky, P, Bray, J, Schoch, G, Phillips, C, Gileadi, C, Soundarajan, M, Yang, X, Elkins, J.M, Gorrec, F, Turnbull, A, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-11 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
2BEL
 
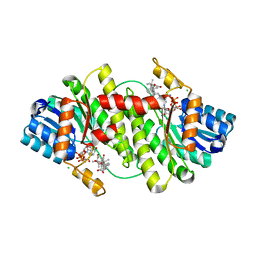 | | Structure of human 11-beta-hydroxysteroid dehydrogenase in complex with NADP and carbenoxolone | | Descriptor: | CARBENOXOLONE, CHLORIDE ION, CORTICOSTEROID 11-BETA-DEHYDROGENASE ISOZYME 1, ... | | Authors: | Kavanagh, K, Wu, X, Svensson, S, Elleby, B, von Delft, F, Debreczeni, J.E, Sharma, S, Bray, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Abrahmsen, L, Oppermann, U. | | Deposit date: | 2004-11-25 | | Release date: | 2004-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The High Resolution Structures of Human, Murine and Guinea Pig 11-Beta-Hydroxysteroid Dehydrogenase Type 1 Reveal Critical Differences in Active Site Architecture
To be Published
|
|
6BAK
 
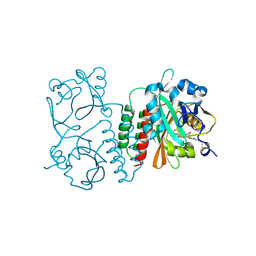 | | The structure of the Stigmatella aurantiaca phytochrome chromophore binding domain T289H mutant | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-13 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
6BAF
 
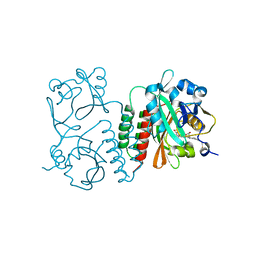 | | Structure of the chromophore binding domain of Stigmatella aurantiaca phytochrome P1, wild-type | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
6BAP
 
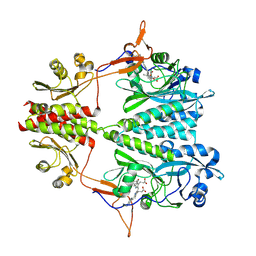 | | Stigmatella aurantiaca bacterial phytochrome PAS-GAF-PHY, T289H mutant | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
6BAY
 
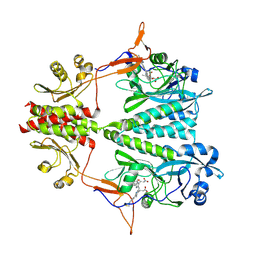 | | Stigmatella aurantiaca bacterial phytochrome P1, PAS-GAF-PHY T289H mutant, room temperature structure | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-16 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
6BAO
 
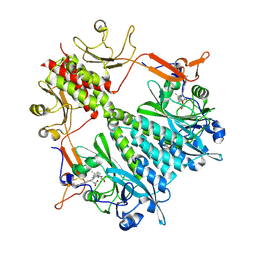 | | Stigmatella aurantiaca phytochrome photosensory core module, wild type | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
5HD3
 
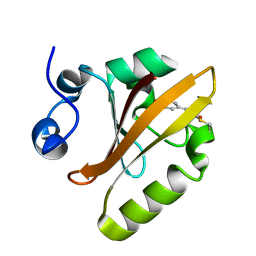 | |
5HDC
 
 | |
5HDD
 
 | |
5HDS
 
 | |
5HD5
 
 | |
