5NNZ
 
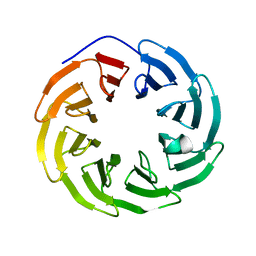 | | Crystal structure of human ODA16 | | Descriptor: | Dynein assembly factor with WDR repeat domains 1 | | Authors: | Lorentzen, E, Taschner, T, Basquin, J. | | Deposit date: | 2017-04-10 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Purification and crystal structure of human ODA16: Implications for ciliary import of outer dynein arms by the intraflagellar transport machinery.
Protein Sci., 29, 2020
|
|
6HYI
 
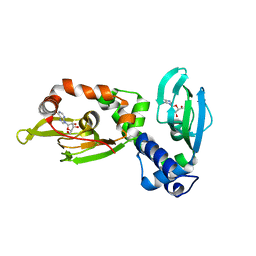 | | Regulatory subunit of a cAMP-independent protein kinase A from Trypanosoma cruzi at 1.4 A resolution in complex with inosine | | Descriptor: | INOSINE, Protein kinase A regulatory subunit | | Authors: | Volpato Santos, Y, Lorentzen, E, Basquin, J, Boshart, M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-11-13 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.39944851 Å) | | Cite: | Purine nucleosides replace cAMP in allosteric regulation of PKA in trypanosomatid pathogens.
Elife, 12, 2024
|
|
6HYQ
 
 | |
2WCZ
 
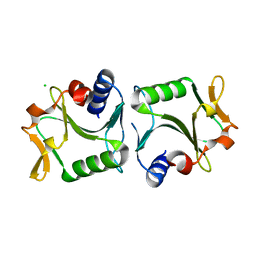 | | 1.6A resolution structure of Archaeoglobus fulgidus Hjc, a Holliday junction resolvase from an archaeal hyperthermophile | | Descriptor: | CHLORIDE ION, HOLLIDAY JUNCTION RESOLVASE | | Authors: | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | Deposit date: | 2009-03-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 1.6 A Resolution Structure of Archaeoglobus Fulgidus Hjc, a Holliday Junction Resolvase from an Archaeal Hyperthermophile
To be Published
|
|
2WIZ
 
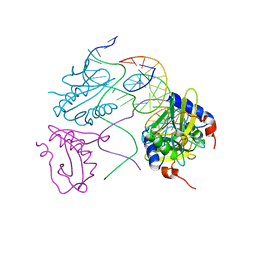 | | Crystal structures of Holliday junction resolvases from Archaeoglobus fulgidus bound to DNA substrate | | Descriptor: | ARCHAEAL HJC, HALF-JUNCTION | | Authors: | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | Deposit date: | 2009-05-18 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structures of Holliday Junction Resolvases from Archaeoglobus Fulgidus Bound to DNA Substrate
To be Published
|
|
2WJ0
 
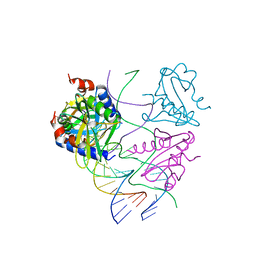 | | Crystal structures of Holliday junction resolvases from Archaeoglobus fulgidus bound to DNA substrate | | Descriptor: | ARCHAEAL HJC, COBALT HEXAMMINE(III), HALF-JUNCTION | | Authors: | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | Deposit date: | 2009-05-18 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of Holliday Junction Resolvases from Archaeoglobus Fulgidus Bound to DNA Substrate
To be Published
|
|
2WIW
 
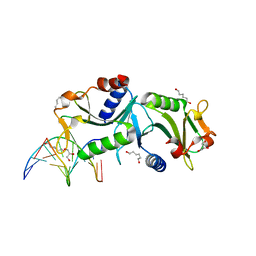 | | Crystal structures of Holliday junction resolvases from Archaeoglobus fulgidus bound to DNA substrate | | Descriptor: | 5'-D(*DC*DG*DG*DA*DT*DA*DT*DC*DC*DGP)-3', GLYCEROL, HEXANE-1,6-DIOL, ... | | Authors: | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | Deposit date: | 2009-05-18 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Holliday Junction Resolvases from Archaeoglobus Fulgidus Bound to DNA Substrate
To be Published
|
|
2WCW
 
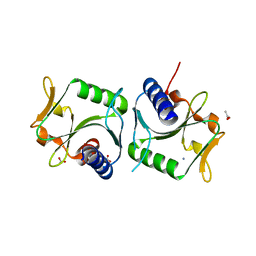 | | 1.6A resolution structure of Archaeoglobus fulgidus Hjc, a Holliday junction resolvase from an archaeal hyperthermophile | | Descriptor: | ACETATE ION, AMMONIUM ION, HJC | | Authors: | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | Deposit date: | 2009-03-17 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | 1.6 A Resolution Structure of Archaeoglobus Fulgidus Hjc, a Holliday Junction Resolvase from an Archaeal Hyperthermophile
To be Published
|
|
2VNU
 
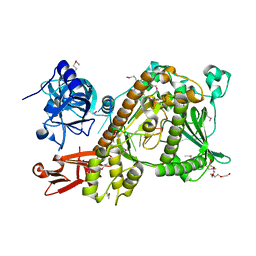 | | Crystal structure of Sc Rrp44 | | Descriptor: | 5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP)-3', EXOSOME COMPLEX EXONUCLEASE RRP44, MAGNESIUM ION, ... | | Authors: | Lorentzen, E, Basquin, J, Conti, E. | | Deposit date: | 2008-02-07 | | Release date: | 2008-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Active Subunit of the Yeast Exosome Core, Rrp44: Diverse Modes of Substrate Recruitment in the Rnase II Nuclease Family
Mol.Cell, 29, 2008
|
|
5OOQ
 
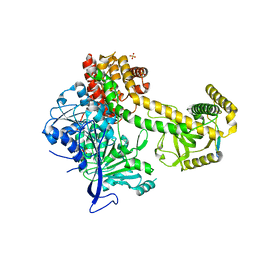 | | Structure of the Mtr4 Nop53 Complex | | Descriptor: | ATP-dependent RNA helicase DOB1, Ribosome biogenesis protein NOP53, SULFATE ION | | Authors: | Falk, S, Basquin, J, Conti, E. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the interaction of the nuclear exosome helicase Mtr4 with the preribosomal protein Nop53.
RNA, 23, 2017
|
|
5NNV
 
 | |
5NMO
 
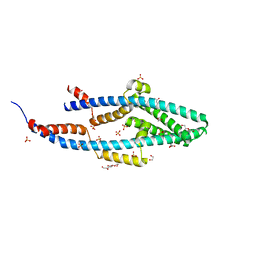 | | Structure of the Bacillus subtilis Smc Joint domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Chromosome partition protein Smc,Chromosome partition protein Smc, ... | | Authors: | Diebold-Durand, M.-L, Basquin, J, Gruber, S. | | Deposit date: | 2017-04-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization.
Mol. Cell, 67, 2017
|
|
3ZGX
 
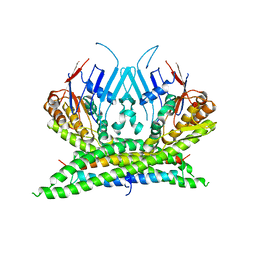 | | Crystal structure of the kleisin-N SMC interface in prokaryotic condensin | | Descriptor: | CHROMOSOME PARTITION PROTEIN SMC, SEGREGATION AND CONDENSATION PROTEIN A | | Authors: | Burmann, F, Shin, H, Basquin, J, Soh, Y, Gimenez, V, Kim, Y, Oh, B, Gruber, S. | | Deposit date: | 2012-12-19 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An Asymmetric Smc-Kleisin Bridge in Prokaryotic Condensin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4BY6
 
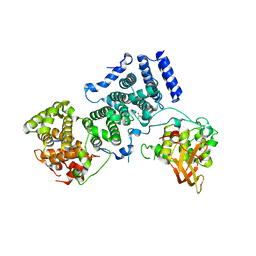 | | Yeast Not1-Not2-Not5 complex | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bhaskar, V, Basquin, J, Conti, E. | | Deposit date: | 2013-07-18 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Structure and RNA-Binding Properties of the not1-not2-not5 Module of the Yeast Ccr4-not Complex
Nat.Struct.Mol.Biol., 20, 2013
|
|
4CT4
 
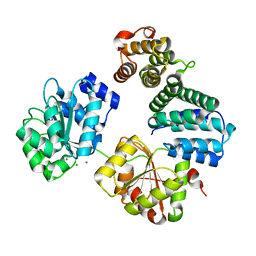 | | CNOT1 MIF4G domain - DDX6 complex | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ozgur, S, Basquin, J, Conti, E. | | Deposit date: | 2014-03-12 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Insights to the Role of the Ccr4- not Complex and Ddx6 ATPase in Microrna Repression.
Mol.Cell, 54, 2014
|
|
4V5W
 
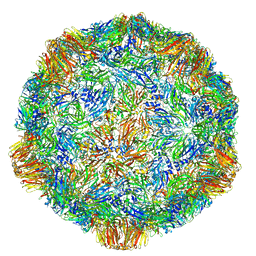 | | Grapevine Fanleaf virus | | Descriptor: | COAT PROTEIN | | Authors: | Schellenberger, P, Demangeat, G, Ritzenthaler, C, Lorber, B, Sauter, C. | | Deposit date: | 2011-05-10 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Exploiting Protein Engineering and Crystal Polymorphism for Successful X-Ray Structure Determination
Cryst.Growth Des., 11, 2011
|
|
5XG2
 
 | |
5XG3
 
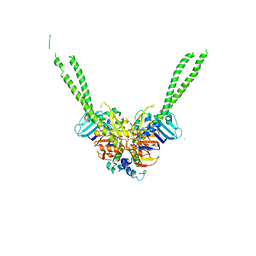 | | Crystal structure of the ATPgS-engaged Smc head domain with an extended coiled coil bound to the C-terminal domain of ScpA derived from Bacillus subtilis | | Descriptor: | COBALT (II) ION, Chromosome partition protein Smc, MAGNESIUM ION, ... | | Authors: | Shin, H.-C, Lee, H, Oh, B.-H. | | Deposit date: | 2017-04-11 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization
Mol. Cell, 67, 2017
|
|
5XNS
 
 | |
5XEI
 
 | |
7Q51
 
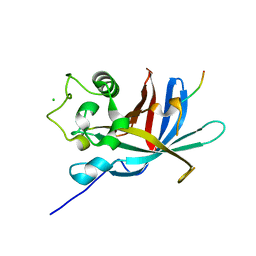 | | yeast Gid10 bound to a Phe/N-peptide | | Descriptor: | CHLORIDE ION, FWLPANLW peptide, Uncharacterized protein YGR066C | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases.
J.Mol.Biol., 434, 2022
|
|
7Q50
 
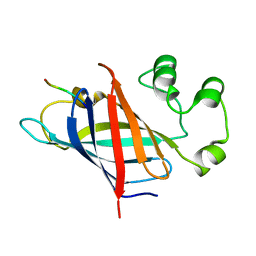 | | human Gid4 bound to a Phe/N-peptide | | Descriptor: | FDVSWFMG peptide, Glucose-induced degradation protein 4 homolog | | Authors: | Chrustowicz, J, Sherpa, D, Loke, M.S, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases.
J.Mol.Biol., 434, 2022
|
|
7Q4Y
 
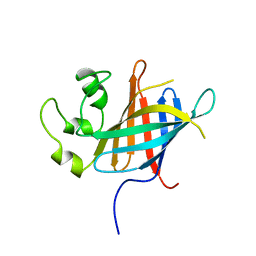 | |
7O6N
 
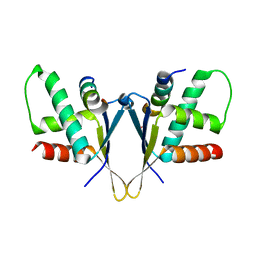 | | Crystal structure of C. elegans ERH-2 PID-3 complex | | Descriptor: | Enhancer of rudimentary homolog 2, FORMIC ACID, Protein pid-3 | | Authors: | Falk, S, Ketting, R.F. | | Deposit date: | 2021-04-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis of PETISCO complex assembly during piRNA biogenesis in C. elegans .
Genes Dev., 35, 2021
|
|
7O6L
 
 | | Crystal structure of C. elegans ERH-2 | | Descriptor: | Enhancer of rudimentary homolog 2 | | Authors: | Falk, S, Ketting, R.F. | | Deposit date: | 2021-04-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of PETISCO complex assembly during piRNA biogenesis in C. elegans .
Genes Dev., 35, 2021
|
|
