5BRB
 
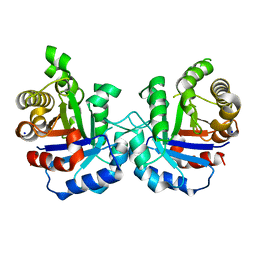 | | Crystal structure of Q64E mutant of Triosephosphate isomerase from Plasmodium falciparum | | Descriptor: | SODIUM ION, Triosephosphate isomerase | | Authors: | Bandyopadhyay, D, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2015-05-30 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Probing the role of highly conserved residues in triosephosphate isomerase - analysis of site specific mutants at positions 64 and 75 in the Plasmodial enzyme
Febs J., 282, 2015
|
|
3KMP
 
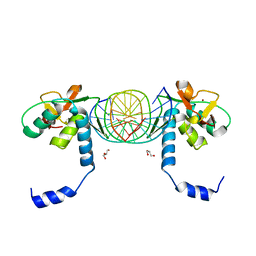 | | Crystal Structure of SMAD1-MH1/DNA complex | | Descriptor: | 5'-D(P*AP*TP*CP*AP*GP*TP*CP*TP*AP*GP*AP*CP*AP*TP*A)-3', 5'-D(P*GP*TP*AP*TP*GP*TP*CP*TP*AP*GP*AP*CP*TP*GP*A)-3', GLYCEROL, ... | | Authors: | Baburajendran, N, Palasingam, P, Narasimhan, K, Jauch, R, Kolatkar, P.R. | | Deposit date: | 2009-11-11 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Smad1 MH1/DNA complex reveals distinctive rearrangements of BMP and TGF-beta effectors.
Nucleic Acids Res., 38, 2010
|
|
3KNG
 
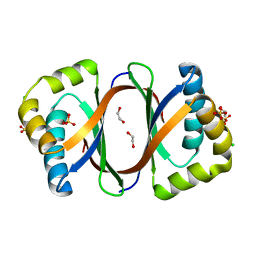 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, determined to 1.9 resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-11-12 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
5JYI
 
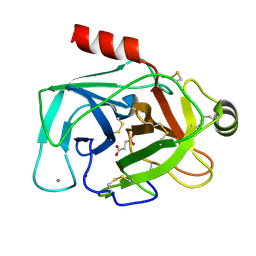 | | Trypsin bound with succinic acid at 1.9A | | Descriptor: | CALCIUM ION, Cationic trypsin, SODIUM ION, ... | | Authors: | Manohar, R, Kutumbarao, N.H.V, KarthiK, L, Malathy, P, Velmurugan, D, Gunasekaran, K. | | Deposit date: | 2016-05-14 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Trypsin bound with succinic acid at 1.9A
To Be Published
|
|
5L95
 
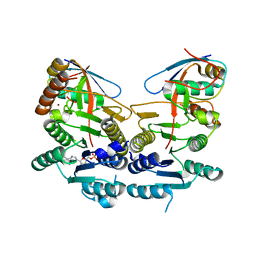 | | Crystal structure of human UBA5 in complex with UFM1 and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ubiquitin-fold modifier 1, Ubiquitin-like modifier-activating enzyme 5, ... | | Authors: | Oweis, W, Padala, P, Wiener, R. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trans-Binding Mechanism of Ubiquitin-like Protein Activation Revealed by a UBA5-UFM1 Complex.
Cell Rep, 16, 2016
|
|
4TOP
 
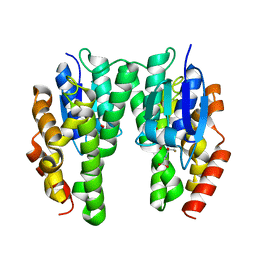 | | Glycine max glutathione transferase | | Descriptor: | 2,4-D inducible glutathione S-transferase, GLUTATHIONE | | Authors: | Axarli, I, Dhavala, P, Papageorgiou, A.C. | | Deposit date: | 2014-06-06 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Comparative analysis of the structural and functional features of two homologous tau class glutathione transferases from Glycine max
To Be Published
|
|
2HD7
 
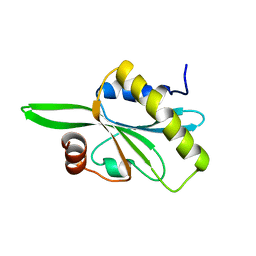 | | Solution structure of C-teminal domain of twinfilin-1. | | Descriptor: | Twinfilin-1 | | Authors: | Hellman, M.H, Paavilainen, V.O, Annila, A, Lappalainen, P, Permi, P.I. | | Deposit date: | 2006-06-20 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis and evolutionary origin of actin filament capping by twinfilin
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2WZL
 
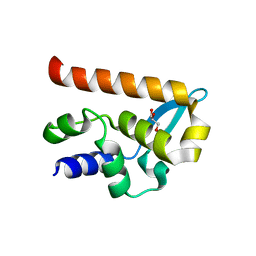 | | The Structure of the N-RNA Binding Domain of the Mokola virus Phosphoprotein | | Descriptor: | GLYCEROL, PHOSPHOPROTEIN | | Authors: | Assenberg, R, Delmas, O, Ren, J, Vidalain, P, Verma, A, Larrous, F, Graham, S, Tangy, F, Grimes, J, Bourhy, H. | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the N-RNA Binding Domain of the Mokola Virus Phosphoprotein
J.Virol., 84, 2010
|
|
3KG1
 
 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, mutant N63A | | Descriptor: | CHLORIDE ION, SnoaB | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
2H4C
 
 | | Structure of Daboiatoxin (heterodimeric PLA2 venom) | | Descriptor: | Phospholipase A2-II, Phospholipase A2-III | | Authors: | Gopalan, G, Thwin, M.M, Gopalakrishnakone, P, Swaminathan, K. | | Deposit date: | 2006-05-24 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and pharmacological comparison of daboiatoxin from Daboia russelli siamensis with viperotoxin F and vipoxin from other vipers.
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
6H77
 
 | | E1 enzyme for ubiquitin like protein activation in complex with UBL | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Soudah, N, Padala, P, Hassouna, F, Mashahreh, B, Lebedev, A.A, Isupov, M.N, Cohen-Kfir, E, Wiener, R. | | Deposit date: | 2018-07-30 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An N-Terminal Extension to UBA5 Adenylation Domain Boosts UFM1 Activation: Isoform-Specific Differences in Ubiquitin-like Protein Activation.
J.Mol.Biol., 431, 2019
|
|
6H78
 
 | | E1 enzyme for ubiquitin like protein activation. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Soudah, N, Padala, P, Hassouna, F, Mashahreh, B, Lebedev, A.A, Isupov, M.N, Cohen-Kfir, E, Wiener, R. | | Deposit date: | 2018-07-30 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An N-Terminal Extension to UBA5 Adenylation Domain Boosts UFM1 Activation: Isoform-Specific Differences in Ubiquitin-like Protein Activation.
J.Mol.Biol., 431, 2019
|
|
2H1P
 
 | | THE THREE-DIMENSIONAL STRUCTURES OF A POLYSACCHARIDE BINDING ANTIBODY TO CRYPTOCOCCUS NEOFORMANS AND ITS COMPLEX WITH A PEPTIDE FROM A PHAGE DISPLAY LIBRARY: IMPLICATIONS FOR THE IDENTIFICATION OF PEPTIDE MIMOTOPES | | Descriptor: | 2H1, PA1 | | Authors: | Young, A.C.M, Valadon, P, Casadevall, A, Scharff, M.D, Sacchettini, J.C. | | Deposit date: | 1997-11-12 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structures of a polysaccharide binding antibody to Cryptococcus neoformans and its complex with a peptide from a phage display library: implications for the identification of peptide mimotopes.
J.Mol.Biol., 274, 1997
|
|
1JY4
 
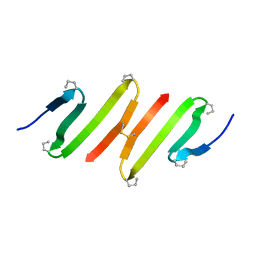 | |
1JY6
 
 | |
5GV4
 
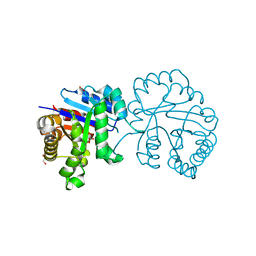 | |
5GIB
 
 | | Succinic acid bound trypsin crystallized as dimer | | Descriptor: | CALCIUM ION, Cationic trypsin, SUCCINIC ACID | | Authors: | Kutumbarao, N.H.V, Manohar, R, KarthiK, L, Malathy, P, Gunasekaran, K, Velmurugan, D. | | Deposit date: | 2016-06-22 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Trypsin bound with Succinic acid
To Be Published
|
|
3DAW
 
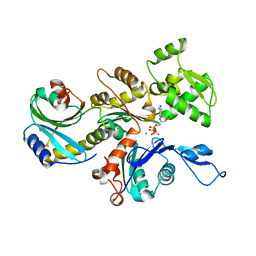 | | Structure of the actin-depolymerizing factor homology domain in complex with actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Paavilainen, V.O, Oksanen, E, Goldman, A, Lappalainen, P. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the actin-depolymerizing factor homology domain in complex with actin
J.Cell Biol., 182, 2008
|
|
5GZP
 
 | |
4KWH
 
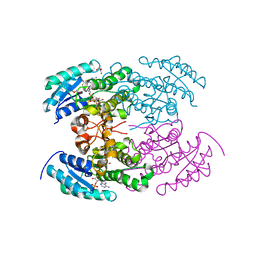 | | The crystal structure of angucycline C-6 ketoreductase LanV with bound NADP | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Paananen, P, Patrikainen, P, Kallio, P, Mantsala, P, Niemi, J, Niiranen, L, Metsa-Ketela, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional analysis of angucycline C-6 ketoreductase LanV involved in landomycin biosynthesis.
Biochemistry, 52, 2013
|
|
4KWI
 
 | | The crystal structure of angucycline C-6 ketoreductase LanV with bound NADP and 11-deoxy-6-oxylandomycinone | | Descriptor: | 1,8-dihydroxy-3-methyltetraphene-6,7,12(5H)-trione, DI(HYDROXYETHYL)ETHER, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Paananen, P, Patrikainen, P, Mantsala, P, Niemi, J, Niiranen, L, Metsa-Ketela, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of angucycline C-6 ketoreductase LanV involved in landomycin biosynthesis.
Biochemistry, 52, 2013
|
|
1CFY
 
 | | YEAST COFILIN, MONOCLINIC CRYSTAL FORM | | Descriptor: | COFILIN | | Authors: | Fedorov, A.A, Lappalainen, P, Fedorov, E.V, Drubin, D.G, Almo, S.C. | | Deposit date: | 1996-11-26 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure determination of yeast cofilin.
Nat.Struct.Biol., 4, 1997
|
|
1HP9
 
 | | kappa-Hefutoxins: a novel Class of Potassium Channel Toxins from Scorpion venom | | Descriptor: | kappa-hefutoxin 1 | | Authors: | Srinivasan, K.N, Sivaraja, V, Huys, I, Sasaki, T, Cheng, B, Kumar, T.K.S, Sato, K, Tytgat, J, Yu, C, Brian Chia, C.S, Ranganathan, S, Bowie, J.H, Kini, R.M, Gopalakrishnakone, P. | | Deposit date: | 2000-12-12 | | Release date: | 2002-08-28 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | kappa-Hefutoxin1, a novel toxin from the scorpion Heterometrus fulvipes with unique structure and function. Importance of the functional diad in potassium channel selectivity.
J.Biol.Chem., 277, 2002
|
|
5IAA
 
 | | Crystal structure of human UBA5 in complex with UFM1 | | Descriptor: | Ubiquitin-fold modifier 1, Ubiquitin-like modifier-activating enzyme 5, ZINC ION | | Authors: | Oweis, W, Padala, P, Wiener, R. | | Deposit date: | 2016-02-21 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Trans-Binding Mechanism of Ubiquitin-like Protein Activation Revealed by a UBA5-UFM1 Complex.
Cell Rep, 16, 2016
|
|
5AGY
 
 | | CRYSTAL STRUCTURE OF A TAU CLASS GST MUTANT FROM GLYCINE | | Descriptor: | 4-NITROPHENYL METHANETHIOL, GLUTATHIONE S-TRANSFERASE, PHOSPHATE ION, ... | | Authors: | Axarli, I, Muleta, A.W, Vlachakis, D, Kossida, S, Kotzia, G, Dhavala, P, Papageorgiou, A.C, Labrou, N.E. | | Deposit date: | 2015-02-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Directed Evolution of Tau Class Glutathione Transferases Reveals a Site that Regulates Catalytic Efficiency and Masks Cooperativity.
Biochem.J., 473, 2016
|
|
