4E0Q
 
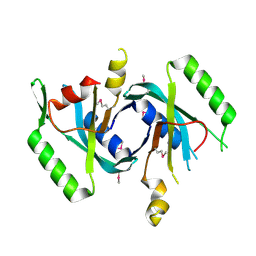 | |
4OCH
 
 | | Apo structure of Smr domain of MutS2 from Deinococcus radiodurans | | Descriptor: | Endonuclease MutS2, GLYCEROL | | Authors: | Zhang, H, Zhao, Y, Xu, Q, Hua, Y.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4001 Å) | | Cite: | Structural and functional studies of MutS2 from Deinococcus radiodurans.
Dna Repair, 21, 2014
|
|
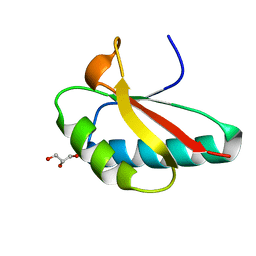
5J5L
 
 | | CRYSTAL STRUCTURE OF AFMP4P IN COMPLEX WITH ARACHIDONIC ACID | | Descriptor: | ARACHIDONIC ACID, Uncharacterized protein | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2016-04-03 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Novel Class Of Virulence Factors In Penicillium Marneffei And Aspergillus Fumigatus Enhances Intracellular Survival In Monocytes By Arachidonic Acid Binding
To Be Published
|
|
4OD6
 
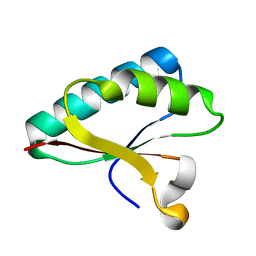 | | Structure of Smr domain of MutS2 from Deinococcus radiodurans, Mn2+ soaked | | Descriptor: | Endonuclease MutS2 | | Authors: | Zhang, H, Zhao, Y, Xu, Q, Hua, Y.J. | | Deposit date: | 2014-01-10 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Structural and functional studies of MutS2 from Deinococcus radiodurans.
Dna Repair, 21, 2014
|
|
5J5K
 
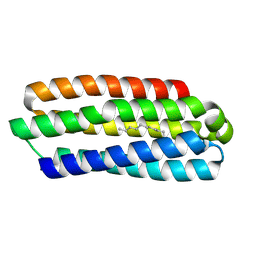 | | CRYSTAL STRUCTURE OF AFMP4P IN COMPLEX WITH PALMITIC ACID | | Descriptor: | PALMITIC ACID, Uncharacterized protein, alpha-D-mannopyranose | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2016-04-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Novel Class Of Virulence Factors In Penicillium Marneffei And Aspergillus Fumigatus Enhances Intracellular Survival In Monocytes By Arachidonic Acid Binding
To Be Published
|
|
4HEG
 
 | | Crystal Structure of HIV-1 protease mutants R8Q complexed with inhibitor GRL-0519 | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, HIV-1 protease | | Authors: | Zhang, H, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2012-10-03 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
4HDB
 
 | | Crystal Structure of HIV-1 protease mutants D30N complexed with inhibitor GRL-0519 | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, HIV-1 Protease, ... | | Authors: | Zhang, H, Wang, Y.-F, Shen, C.H, Agniswamy, J, Weber, I.T. | | Deposit date: | 2012-10-02 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
4HDF
 
 | | Crystal Structure of HIV-1 protease mutants V82A complexed with inhibitor GRL-0519 | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, HIV-1 Protease | | Authors: | Zhang, H, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2012-10-02 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
5ZGE
 
 | | Crystal structure of NDM-1 at pH5.5 (Bis-Tris) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, HYDROXIDE ION, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
4JUR
 
 | |
4KVG
 
 | | Crystal structure of RIAM RA-PH domains in complex with GTP bound Rap1 | | Descriptor: | 1,2-ETHANEDIOL, Amyloid beta A4 precursor protein-binding family B member 1-interacting protein, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, H, Chang, Y.E, Brennan, M.L, Wu, J. | | Deposit date: | 2013-05-22 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of Rap1 in complex with RIAM reveals specificity determinants and recruitment mechanism.
J Mol Cell Biol, 6, 2014
|
|
4KZ2
 
 | | Crystal Structure of phi29 pRNA 3WJ Core | | Descriptor: | MANGANESE (II) ION, phi29 pRNA 3WJ core RNA 16 mer, phi29 pRNA 3WJ core RNA 18 mer, ... | | Authors: | Zhang, H, Endrizzi, J.A, Shu, Y, Haque, F, Sauter, C, Guo, P, Chi, Y.-I. | | Deposit date: | 2013-05-29 | | Release date: | 2013-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA.
Rna, 19, 2013
|
|
5GRM
 
 | | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP) | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-11 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP)
To Be Published
|
|
3RAJ
 
 | |
3OZO
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with NGT | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, N-acetylglucosaminidase | | Authors: | Zhang, H, Liu, T, Liu, F, Shen, X, Yang, Q. | | Deposit date: | 2010-09-27 | | Release date: | 2011-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with NGT
To be Published
|
|
8HS8
 
 | |
5ZGU
 
 | |
5ZH1
 
 | |
5ZGR
 
 | | Crystal structure of NDM-1 at pH7.3 (HEPES) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-10 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
5ZGV
 
 | |
5ZGY
 
 | |
5ZGW
 
 | |
5ZGQ
 
 | | Crystal structure of NDM-1 at pH7.5 (Tris-HCl, (NH4)2SO4) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, HYDROXIDE ION, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-10 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
5ZGZ
 
 | |
6B57
 
 | | tudor in complex with ligand | | Descriptor: | Tudor and KH domain-containing protein, UNKNOWN ATOM OR ION | | Authors: | Zhang, H, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-28 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for arginine methylation-independent recognition of PIWIL1 by TDRD2.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
