5AE1
 
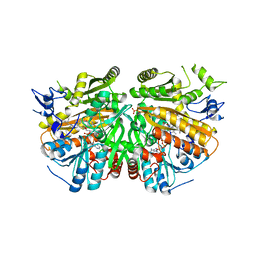 | | Ether Lipid-Generating Enzyme AGPS in complex with inhibitor ZINC69435460 | | Descriptor: | (3-(2-FLUOROPHENYL)-N-(1-(2-OXO-2,3-DIHYDRO-1H-BENZO[D]IMIDAZOL-5-YL)ETHYL)BUTANAMIDE), ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE, PEROXISOMAL, ... | | Authors: | Piano, V, Benjamin, D.I, Valente, S, Nenci, S, Marrocco, B, Mai, A, Aliverti, A, Nomura, D.K, Mattevi, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Inhibitors for the Ether Lipid-Generating Enzyme Agps as Anti-Cancer Agents.
Acs Chem.Biol., 10, 2015
|
|
5ADZ
 
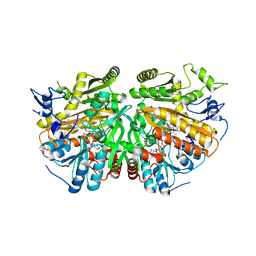 | | Ether Lipid-Generating Enzyme AGPS in complex with inhibitor 1a | | Descriptor: | (3S)-3-(2-fluorophenyl)-N-((2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)methyl)butanamide), ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE, PEROXISOMAL, ... | | Authors: | Piano, V, Benjamin, D.I, Valente, S, Nenci, S, Marrocco, B, Mai, A, Aliverti, A, Nomura, D.K, Mattevi, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Inhibitors for the Ether Lipid-Generating Enzyme Agps as Anti-Cancer Agents.
Acs Chem.Biol., 10, 2015
|
|
6ZLJ
 
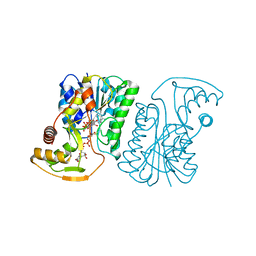 | | Crystal Structure of UDP-Glucuronic acid 4-epimerase Y149F mutant from Bacillus cereus in complex with UDP-4-DEOXY-4-FLUORO-Glucuronic acid and NAD | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{R})-6-[[[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidany l)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3-fluoranyl-4,5-bis(oxidanyl)oxane-2-carboxylic acid, Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Iacovino, L.G, Mattevi, A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
6ZL6
 
 | |
6ZLD
 
 | | Crystal Structure of UDP-Glucuronic acid 4-epimerase from Bacillus cereus in complex with UDP-Glucuronic acid and NAD | | Descriptor: | Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Iacovino, L.G, Savino, S, Mattevi, A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
6ZLL
 
 | | Crystal Structure of UDP-Glucuronic acid 4-epimerase from Bacillus cereus in complex with UDP-Galacturonic acid and NAD | | Descriptor: | (2S,3R,4S,5R,6R)-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4,5-trihydroxy-oxane-2-carboxylic acid, Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Iacovino, L.G, Savino, S, Mattevi, A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
2C7G
 
 | | FprA from Mycobacterium tuberculosis: His57Gln mutant | | Descriptor: | 4-OXO-NICOTINAMIDE-ADENINE DINUCLEOTIDE PHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH-FERREDOXIN REDUCTASE FPRA, ... | | Authors: | Pennati, A, Razeto, A, De Rosa, M, Pandini, V, Vanoni, M.A, Aliverti, A, Mattevi, A, Coda, A, Zanetti, G. | | Deposit date: | 2005-11-24 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the His57-Glu214 Ionic Couple Located in the Active Site of Mycobacterium Tuberculosis Fpra.
Biochemistry, 45, 2006
|
|
8S7S
 
 | |
8S7Q
 
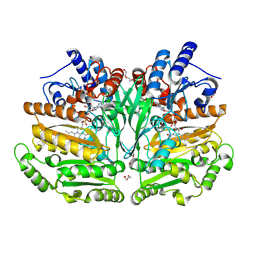 | |
8S7R
 
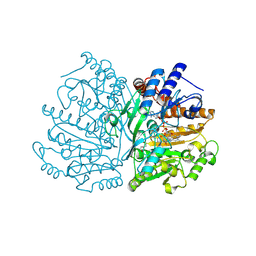 | |
8S7U
 
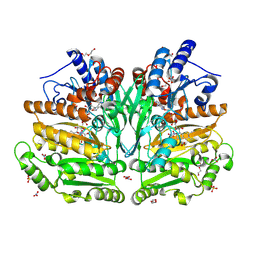 | |
8S7N
 
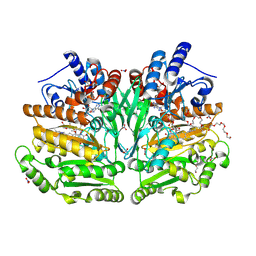 | |
8S7P
 
 | |
8S7T
 
 | |
8S7W
 
 | |
2JBR
 
 | |
2JBS
 
 | |
2JBT
 
 | |
8QNR
 
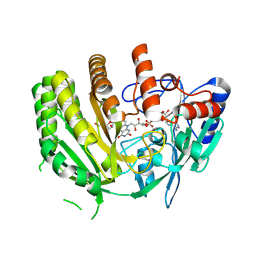 | | Crystal structure of ancestral L-galactono-1,4-lactone dehydrogenase: G413N variant in complex with L-gulono-1,4-lactone | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-galactono-1,4-lactone dehydrogenase, L-gulono-1,4-lactone | | Authors: | Boverio, A, Mattevi, A. | | Deposit date: | 2023-09-27 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure, mechanism, and evolution of the last step in vitamin C biosynthesis.
Nat Commun, 15, 2024
|
|
8QMY
 
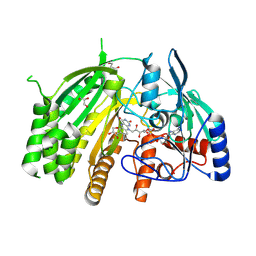 | | Crystal structure of ancestral L-galactono-1,4-lactone dehydrogenase | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Boverio, A, Mattevi, A. | | Deposit date: | 2023-09-25 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure, mechanism, and evolution of the last step in vitamin C biosynthesis.
Nat Commun, 15, 2024
|
|
8QNB
 
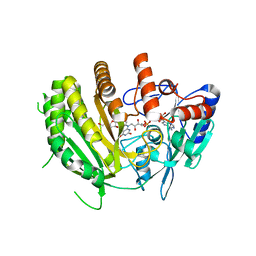 | | Crystal structure of ancestral L-galactono-1,4-lactone dehydrogenase: in complex with L-galactono-1,4-lactone | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-galactono-1,4-lactone, L-galactono-1,4-lactone dehydrogenase | | Authors: | Boverio, A, Mattevi, A. | | Deposit date: | 2023-09-26 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure, mechanism, and evolution of the last step in vitamin C biosynthesis.
Nat Commun, 15, 2024
|
|
8QNC
 
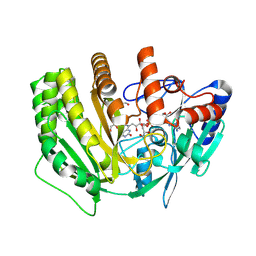 | | Crystal structure of ancestral L-galactono-1,4-lactone dehydrogenase: A113G variant | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Boverio, A, Mattevi, A. | | Deposit date: | 2023-09-26 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure, mechanism, and evolution of the last step in vitamin C biosynthesis.
Nat Commun, 15, 2024
|
|
6ZLA
 
 | |
6ZLK
 
 | | Equilibrium Structure of UDP-Glucuronic acid 4-epimerase from Bacillus cereus in complex with UDP-Glucuronic acid/UDP-Galacturonic acid and NAD | | Descriptor: | (2S,3R,4S,5R,6R)-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4,5-trihydroxy-oxane-2-carboxylic acid, Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Iacovino, L.G, Mattevi, A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
8ULC
 
 | | LSD1-CoREST in complex with T15, long soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, methyl 3-{(1R,3S,3aS,13R)-8-[(2S,3S,4R)-5-{[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-2,3,4-trihydroxypentyl]-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,4,5,6,8-hexahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-3-yl}benzoate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-10-16 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
To Be Published
|
|
