4LT8
 
 | | Crystal Structure of tRNA Proline (CGG) Bound to Codon CCC-G on the Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-07-23 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.14000177 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4LNT
 
 | | Crystal Structure of tRNA Proline (CGG) Bound to Codon CCC-U on the Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-07-12 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4LSK
 
 | | Crystal Structure of tRNA Proline (CGG) Bound to Codon CCG-G on the Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-07-22 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.48000622 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MCT
 
 | |
4MCX
 
 | |
4PX8
 
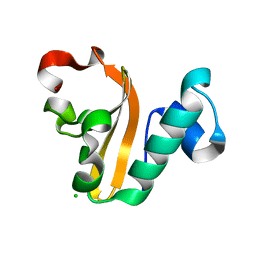 | | Structure of P. vulgaris HigB toxin | | Descriptor: | CHLORIDE ION, Killer protein | | Authors: | Schureck, M.A, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2014-03-22 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Defining the mRNA recognition signature of a bacterial toxin protein.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4P6F
 
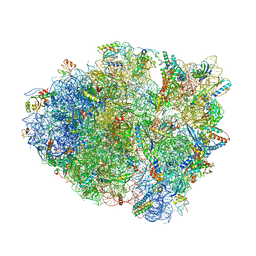 | |
4P70
 
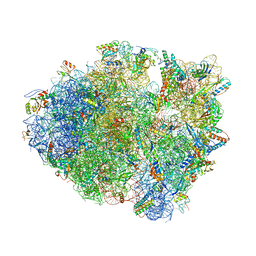 | | Crystal Structure of Unmodified tRNA Proline (CGG) Bound to Codon CCG on the Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2014-03-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4OX9
 
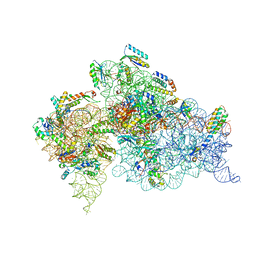 | | Crystal structure of the aminoglycoside resistance methyltransferase NpmA bound to the 30S ribosomal subunit | | Descriptor: | 16S rRNA, 16S rRNA (adenine(1408)-N(1))-methyltransferase, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Conn, G.L, Dunham, C.M. | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.8035 Å) | | Cite: | Molecular recognition and modification of the 30S ribosome by the aminoglycoside-resistance methyltransferase NpmA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TUA
 
 | |
4TUC
 
 | |
4TUE
 
 | |
4TUD
 
 | |
4Q2U
 
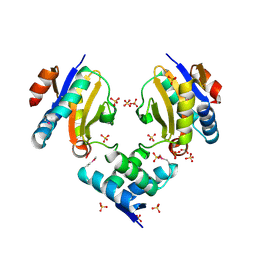 | |
4TUB
 
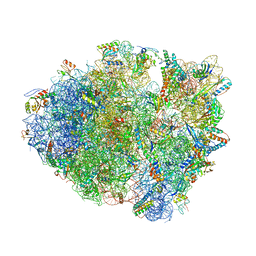 | |
6CF1
 
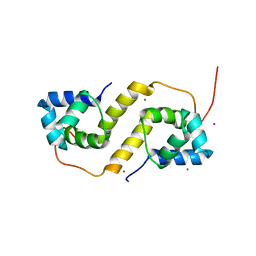 | | Proteus vulgaris HigA antitoxin structure | | Descriptor: | Antitoxin HigA, POTASSIUM ION | | Authors: | Schureck, M.A, Hoffer, E.D, Ei Cho, S, Dunham, C.M. | | Deposit date: | 2018-02-13 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of transcriptional regulation by the HigA antitoxin.
Mol.Microbiol., 111, 2019
|
|
6CHV
 
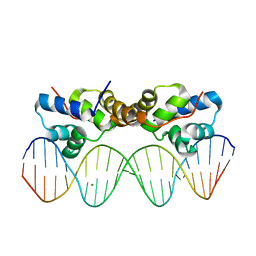 | | Proteus vulgaris HigA antitoxin bound to DNA | | Descriptor: | Antitoxin HigA, MAGNESIUM ION, pHigCryst3, ... | | Authors: | Schureck, M.A, Hoffer, E.D, Onuoha, N, Dunham, C.M. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of transcriptional regulation by the HigA antitoxin.
Mol.Microbiol., 111, 2019
|
|
6BZ8
 
 | | Thermus thermophilus 70S containing 16S G347U point mutation and near-cognate ASL Leucine in A site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2017-12-22 | | Release date: | 2018-11-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | Ribosomal ambiguity (ram) mutations promote the open (off) to closed (on) transition and thereby increase miscoding.
Nucleic Acids Res., 47, 2019
|
|
6BZ6
 
 | | Thermus thermophilus 70S complex containing 16S G347U ram mutation and empty A site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2017-12-22 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Ribosomal ambiguity (ram) mutations promote the open (off) to closed (on) transition and thereby increase miscoding.
Nucleic Acids Res., 47, 2019
|
|
6BUW
 
 | | Thermus thermophilus 70S complex containing 16S G299A ram mutation and empty A site. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2017-12-11 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Ribosomal ambiguity (ram) mutations promote the open (off) to closed (on) transition and thereby increase miscoding.
Nucleic Acids Res., 47, 2019
|
|
6BZ7
 
 | | Thermus thermophilus 70S containing 16S G299A point mutation and near-cognate ASL Leucine in A site. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2017-12-22 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Ribosomal ambiguity (ram) mutations promote the open (off) to closed (on) transition and thereby increase miscoding.
Nucleic Acids Res., 47, 2019
|
|
6PQB
 
 | |
2CMN
 
 | | A Proximal Arginine Residue in the Switching Mechanism of the FixL Oxygen Sensor | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SENSOR PROTEIN FIXL | | Authors: | Gilles-Gonzalez, M.-A, Caceres, A.I, Silva Sousa, E.H, Tomchick, D.R, Brautigam, C.A, Gonzalez, C, Machius, M. | | Deposit date: | 2006-05-11 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Proximal Arginine R206 Participates in Switching of the Bradyrhizobium Japonicum Fixl Oxygen Sensor
J.Mol.Biol., 360, 2006
|
|
4V5K
 
 | | Structure of cytotoxic domain of colicin E3 bound to the 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Meenan, N.A.G, Sharma, A, Kelley, A.C, Kleanthous, C, Ramakrishnan, V. | | Deposit date: | 2010-05-29 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for 16S Ribosomal RNA Cleavage by the Cytotoxic Domain of Colicin E3.
Nat.Struct.Mol.Biol., 17, 2010
|
|
9E0N
 
 | | M. smegmatis unmethylated 70S ribosome structure | | Descriptor: | 16S rRNA, 23S rRNA, 50S ribosomal protein L36, ... | | Authors: | Nandi, S, Conn, G.L. | | Deposit date: | 2024-10-18 | | Release date: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Distant ribose 2'-O-methylation of 23S rRNA helix 69 pre-orders the capreomycin drug binding pocket at the ribosome subunit interface.
Nucleic Acids Res., 53, 2025
|
|
