8A4L
 
 | | Lipidic alpha-synuclein fibril - polymorph L2A | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-06-12 | | Release date: | 2022-08-17 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
8ADW
 
 | | Lipidic alpha-synuclein fibril - polymorph L1C | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-07-11 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
8ADU
 
 | | Lipidic alpha-synuclein fibril - polymorph L1A | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-07-11 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
2MKC
 
 | | Cooperative Structure of the Heterotrimeric pre-mRNA Retention and Splicing Complex | | Descriptor: | Pre-mRNA leakage protein 1, Pre-mRNA-splicing factor CWC26, U2 snRNP component IST3 | | Authors: | Wysoczanski, P, Schneider, C, Xiang, S, Munari, F, Trowitzsch, S, Wahl, M.C, Luhrmann, R, Becker, S, Zweckstetter, M. | | Deposit date: | 2014-02-04 | | Release date: | 2014-09-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cooperative structure of the heterotrimeric pre-mRNA retention and splicing complex.
Nat.Struct.Mol.Biol., 21, 2014
|
|
2N64
 
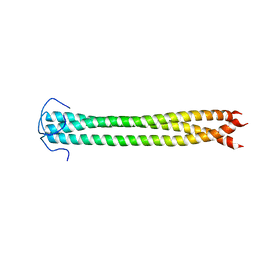 | |
2JK4
 
 | | Structure of the human voltage-dependent anion channel | | Descriptor: | VOLTAGE-DEPENDENT ANION-SELECTIVE CHANNEL PROTEIN 1 | | Authors: | Bayrhuber, M, Meins, T, Habeck, M, Becker, S, Giller, K, Villinger, S, Vonrhein, C, Griesinger, C, Zweckstetter, M, Zeth, K. | | Deposit date: | 2008-08-15 | | Release date: | 2008-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structure of the Human Voltage-Dependent Anion Channel.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2J80
 
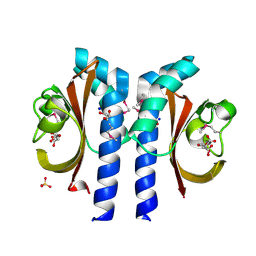 | | Structure of Citrate-bound Periplasmic Domain of Sensor Histidine Kinase CitA | | Descriptor: | CITRATE ANION, GLYCEROL, SENSOR KINASE CITA, ... | | Authors: | Sevvana, M, Vijayan, V, Zweckstetter, M, Reinelt, S, Madden, D.R, Sheldrick, G.M, Bott, M, Griesinger, C, Becker, S. | | Deposit date: | 2006-10-18 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Ligand-Induced Switch in the Periplasmic Domain of Sensor Histidine Kinase Cita.
J.Mol.Biol., 377, 2008
|
|
2MNH
 
 | | Refined structure of outer membrane protein x in nanodisc by measuring residual dipolar couplings | | Descriptor: | Outer membrane protein X | | Authors: | Bibow, S, Carneiro, M.G, Sabo, T.M, Schwiegk, C, Becker, S, Riek, R, Lee, D. | | Deposit date: | 2014-04-05 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Measuring membrane protein bond orientations in nanodiscs via residual dipolar couplings.
Protein Sci., 23, 2014
|
|
2N7H
 
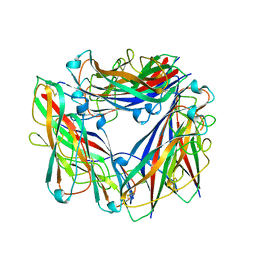 | | Hybrid structure of the Type 1 Pilus of Uropathogenic E.coli | | Descriptor: | FimA | | Authors: | Habenstein, B, Loquet, A, Giller, K, Vasa, S, Becker, S, Habeck, M, Lange, A. | | Deposit date: | 2015-09-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-20 | | Method: | SOLID-STATE NMR | | Cite: | Hybrid Structure of the Type 1 Pilus of Uropathogenic Escherichia coli.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2MSG
 
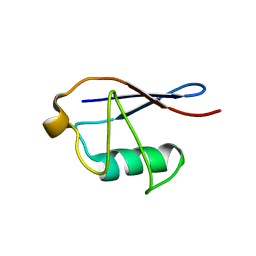 | | Solid-state NMR structure of ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Lakomek, N, Habenstein, B, Loquet, A, Shi, C, Giller, K, Wolff, S, Becker, S, Fasshuber, H, Lange, A. | | Deposit date: | 2014-08-04 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structural heterogeneity in microcrystalline ubiquitin studied by solid-state NMR.
Protein Sci., 24, 2015
|
|
2KOX
 
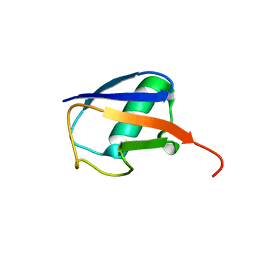 | | NMR residual dipolar couplings identify long range correlated motions in the backbone of the protein ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Fenwick, R.B, Richter, B, Lee, D, Walter, K.F.A, Milovanovic, D, Becker, S, Lakomek, N.A, Griesinger, C, Salvatella, X. | | Deposit date: | 2009-10-02 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Weak Long-Range Correlated Motions in a Surface Patch of Ubiquitin Involved in Molecular Recognition
J.Am.Chem.Soc., 2011
|
|
2MME
 
 | | Hybrid structure of the Shigella flexneri MxiH Type three secretion system needle | | Descriptor: | MxiH | | Authors: | Demers, J.P, Habenstein, B, Loquet, A, Vasa, S.K, Becker, S, Baker, D, Lange, A, Sgourakis, N.G. | | Deposit date: | 2014-03-14 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.7 Å), SOLID-STATE NMR | | Cite: | High-resolution structure of the Shigella type-III secretion needle by solid-state NMR and cryo-electron microscopy.
Nat Commun, 5, 2014
|
|
2MF7
 
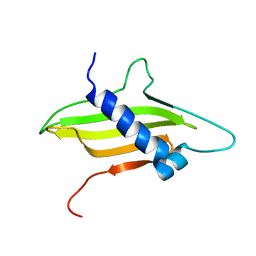 | | Solution structure of the ims domain of the mitochondrial import protein TIM21 from S. cerevisiae | | Descriptor: | Mitochondrial import inner membrane translocase subunit TIM21 | | Authors: | Bajaj, R, Jaremko, L, Jaremko, M, Becker, S, Zweckstetter, M. | | Deposit date: | 2013-10-07 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis of the Dynamic Structure of the TIM23 Complex in the Mitochondrial Intermembrane Space.
Structure, 22, 2014
|
|
2LPZ
 
 | | Atomic model of the Type-III Secretion System Needle | | Descriptor: | Protein prgI | | Authors: | Loquet, A, Sgourakis, N.G, Gupta, R, Giller, K, Riedel, D, Goosmann, C, Griesinger, C, Kolbe, M.G, Baker, D, Becker, S, Lange, A. | | Deposit date: | 2012-02-21 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic model of the type III secretion system needle.
Nature, 486, 2012
|
|
2M97
 
 | |
2J82
 
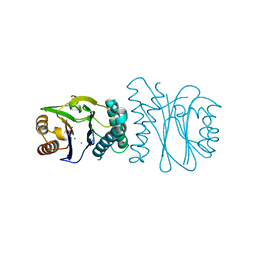 | | Structural analysis of the PP2C Family Phosphatase tPphA from Thermosynechococcus elongatus | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PROTEIN SERINE-THREONINE PHOSPHATASE | | Authors: | Schlicker, C, Kloft, N, Forchhammer, K, Becker, S. | | Deposit date: | 2006-10-18 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural Analysis of the Pp2C Phosphatase Tppha from Thermosynechococcus Elongatus: A Flexible Flap Subdomain Controls Access to the Catalytic Site.
J.Mol.Biol., 376, 2008
|
|
2LYQ
 
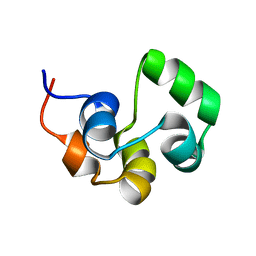 | | NOE-based 3D structure of the monomeric intermediate of CylR2 at 262K (-11 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2MGY
 
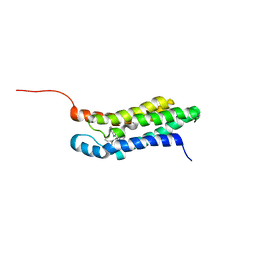 | | Solution structure of the mitochondrial translocator protein (TSPO) in complex with its high-affinity ligand PK11195 | | Descriptor: | N-[(2R)-butan-2-yl]-1-(2-chlorophenyl)-N-methylisoquinoline-3-carboxamide, Translocator protein | | Authors: | Jaremko, M, Jaremko, L, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the mitochondrial translocator protein in complex with a diagnostic ligand.
Science, 343, 2014
|
|
2K3H
 
 | | Structural determinants for Ca2+ and PIP2 binding by the C2A domain of rabphilin-3A | | Descriptor: | CALCIUM ION, Rabphilin-3A | | Authors: | Coudevylle, N, Montaville, P, Leonov, A, Zweckstetter, M, Becker, S. | | Deposit date: | 2008-05-08 | | Release date: | 2008-10-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Determinants for Ca2+ and Phosphatidylinositol 4,5-Bisphosphate Binding by the C2A Domain of Rabphilin-3A.
J.Biol.Chem., 283, 2008
|
|
2K39
 
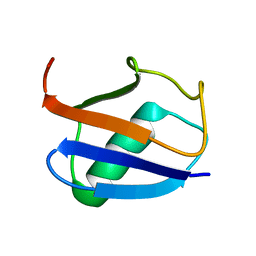 | | Recognition dynamics up to microseconds revealed from RDC derived ubiquitin ensemble in solution | | Descriptor: | Ubiquitin | | Authors: | Lange, O.F, Lakomek, N.A, Fares, C, Schroder, G, Walter, K, Becker, S, Meiler, J, Grubmuller, H, Griesinger, C, de Groot, B.L. | | Deposit date: | 2008-04-25 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution.
Science, 320, 2008
|
|
2LYJ
 
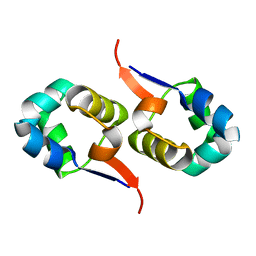 | | NOE-based 3D structure of the CylR2 homodimer at 298K | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Giller, K, Becker, S, Zweckstetter, M, Schwieters, C.D. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYK
 
 | | NOE-based 3D structure of the CylR2 homodimer at 270K (-3 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2MY2
 
 | |
2LYS
 
 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 257K (-16 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYR
 
 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 259K (-14 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
