1V3F
 
 | |
1V63
 
 | | Solution structure of the 6th HMG box of mouse UBF1 | | Descriptor: | Nucleolar transcription factor 1 | | Authors: | Sato, M, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-27 | | Release date: | 2004-05-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 6th HMG box of mouse UBF1
To be Published
|
|
2RRT
 
 | |
1UHM
 
 | | Solution structure of the globular domain of linker histone homolog Hho1p from S. cerevisiae | | Descriptor: | Histone H1 | | Authors: | Ono, K, Kusano, O, Shimotakahara, S, Shimizu, M, Yamazaki, T, Shindo, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-05 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The linker histone homolog Hho1p from Saccharomyces cerevisiae represents a winged helix-turn-helix fold as determined by NMR spectroscopy.
Nucleic Acids Res., 31, 2003
|
|
1UHS
 
 | | Solution structure of mouse homeodomain-only protein HOP | | Descriptor: | homeodomain only protein | | Authors: | Saito, K, Koshiba, S, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-10 | | Release date: | 2004-07-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mouse homeodomain-only protein HOP
To be Published
|
|
1UI1
 
 | | Crystal Structure Of Uracil-DNA Glycosylase From Thermus Thermophilus HB8 | | Descriptor: | IRON/SULFUR CLUSTER, Uracil-DNA Glycosylase | | Authors: | Hoseki, J, Okamoto, A, Masui, R, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-14 | | Release date: | 2003-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Family 4 Uracil-DNA Glycosylase from Thermus thermophilus HB8
J.Mol.Biol., 333, 2003
|
|
1UI0
 
 | | Crystal Structure Of Uracil-DNA Glycosylase From Thermus Thermophilus HB8 | | Descriptor: | IRON/SULFUR CLUSTER, SULFATE ION, URACIL, ... | | Authors: | Hoseki, J, Okamoto, A, Masui, R, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-14 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Family 4 Uracil-DNA Glycosylase from Thermus thermophilus HB8
J.Mol.Biol., 333, 2003
|
|
1UHR
 
 | | Solution structure of the SWIB domain of mouse BRG1-associated factor 60a | | Descriptor: | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily D member 1 | | Authors: | Yamada, K, Saito, K, Nameki, N, Inoue, M, Koshiba, S, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-09 | | Release date: | 2004-08-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SWIB domain of mouse BRG1-associated factor 60a
To be Published
|
|
1UIR
 
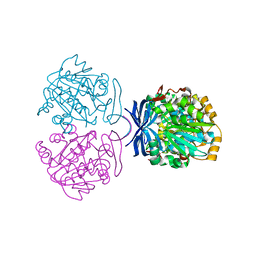 | | Crystal Structure of Polyamine Aminopropyltransfease from Thermus thermophilus | | Descriptor: | Polyamine Aminopropyltransferase | | Authors: | Ganbe, T, Ohnuma, M, Sato, T, Kumasaka, T, Oshima, T, Tanaka, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-18 | | Release date: | 2003-08-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and enzymatic properties of a triamine/agmatine aminopropyltransferase from Thermus thermophilus
J.Mol.Biol., 408, 2011
|
|
1UJP
 
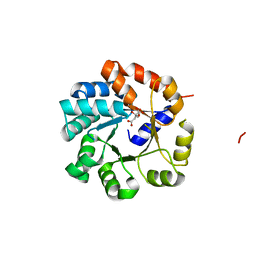 | | Crystal Structure of Tryptophan Synthase A-Subunit From Thermus thermophilus HB8 | | Descriptor: | CITRIC ACID, Tryptophan synthase alpha chain | | Authors: | Asada, Y, Yokoyama, S, Kuramitsu, S, Miyano, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-08 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Stabilization mechanism of the tryptophan synthase alpha-subunit from Thermus thermophilus HB8: X-ray crystallographic analysis and calorimetry.
J.Biochem.(Tokyo), 138, 2005
|
|
2P9X
 
 | |
1UIL
 
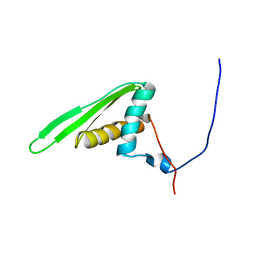 | | Double-stranded RNA-binding motif of Hypothetical protein BAB28848 | | Descriptor: | Double-stranded RNA-binding motif | | Authors: | Nagata, T, Muto, Y, Hayashi, F, Hamana, H, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-17 | | Release date: | 2004-11-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of Double-stranded RNA-binding motif of Hypothetical protein BAB28848
To be Published
|
|
1UJS
 
 | |
2P5D
 
 | |
1V5Q
 
 | | Solution Structure of the PDZ Domain from Mouse Glutamate Receptor Interacting Protein 1A-L (GRIP1) Homolog | | Descriptor: | Glutamate Receptor Interacting Protein 1A-L Homolog | | Authors: | Sato, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the PDZ Domain from Mouse Glutamate Receptor Interacting Protein 1A-L (GRIP1) Homolog
To be Published
|
|
1V80
 
 | |
1V3V
 
 | | Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase complexed with NADP and 15-oxo-PGE2 | | Descriptor: | (5E,13E)-11-HYDROXY-9,15-DIOXOPROSTA-5,13-DIEN-1-OIC ACID, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hori, T, Yokomizo, T, Ago, H, Sugahara, M, Ueno, G, Yamamoto, M, Kumasaka, T, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-06 | | Release date: | 2004-07-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop
J.Biol.Chem., 279, 2004
|
|
1V61
 
 | | Solution Structure of the Pleckstrin Homology Domain of alpha-Pix | | Descriptor: | Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 | | Authors: | Nakanishi, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-26 | | Release date: | 2004-05-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Pleckstrin Homology Domain of alpha-Pix
To be Published
|
|
1V81
 
 | |
2PD2
 
 | | Crystal structure of (ST0148) conserved hypothetical from Sulfolobus Tokodaii Strain7 | | Descriptor: | Hypothetical protein ST0148 | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Rafi, Z.A, Sekar, K, Agari, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-31 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of (ST0148) conserved hypothetical from Sulfolobus Tokodaii Strain7
To be Published
|
|
1V5U
 
 | | Solution Structure of the C-terminal Pleckstrin Homology Domain of Sbf1 from Mouse | | Descriptor: | SET binding factor 1 | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Pleckstrin Homology Domain of Sbf1 from Mouse
To be Published
|
|
1V6T
 
 | |
1V3R
 
 | | Crystal structure of TT1020 from Thermus thermophilus HB8 | | Descriptor: | Nitrogen regulatory protein P-II | | Authors: | Wang, H, Sakai, H, Takemoto-Hori, C, Kaminishi, T, Yamaguchi, H, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-05 | | Release date: | 2004-11-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the signal transducing protein GlnK from Thermus thermophilus HB8.
J.Struct.Biol., 149, 2005
|
|
1V5W
 
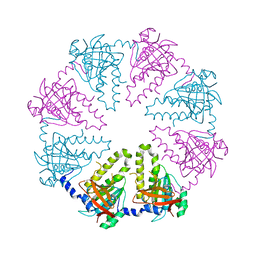 | | Crystal structure of the human Dmc1 protein | | Descriptor: | Meiotic recombination protein DMC1/LIM15 homolog | | Authors: | Kinebuchi, T, Kagawa, W, Enomoto, R, Ikawa, S, Shibata, T, Kurumizaka, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-26 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein dmc1
Mol.Cell, 14, 2004
|
|
1V3T
 
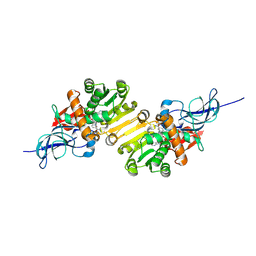 | | Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, leukotriene b4 12-hydroxydehydrogenase/prostaglandin 15-keto reductase | | Authors: | Hori, T, Yokomizo, T, Ago, H, Sugahara, M, Ueno, G, Yamamoto, M, Kumasaka, T, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-05 | | Release date: | 2004-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop
J.Biol.Chem., 279, 2004
|
|
