7KK3
 
 | | Structure of the catalytic domain of PARP1 in complex with talazoparib | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KKZ
 
 | | Crystal structure of mouse anti-HIV potent neutralizing antibody M4H2K1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Heavy chain of Fab fragment of mouse monoclonal antibody M4H2K1, ... | | Authors: | Kumar, S, Wilson, I.A. | | Deposit date: | 2020-10-28 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Neutralizing Antibodies Induced by First-Generation gp41-Stabilized HIV-1 Envelope Trimers and Nanoparticles.
Mbio, 12, 2021
|
|
7KMD
 
 | |
7KLC
 
 | |
7KNW
 
 | | Crystal structure of SND1 in complex with C-26-A2 | | Descriptor: | 5-chloro-2-methoxy-N-([1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1 | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
7KNX
 
 | | Crystal structure of SND1 in complex with C-26-A6 | | Descriptor: | 5-chloro-2-methoxy-N-(2-methyl[1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1, ... | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
6K9N
 
 | | Rice_OTUB_like_catalytic domain | | Descriptor: | Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
6ILC
 
 | |
6IEH
 
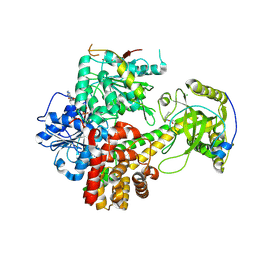 | | Crystal structures of the hMTR4-NRDE2 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Exosome RNA helicase MTR4, ... | | Authors: | Chen, J.Y, Yun, C.H. | | Deposit date: | 2018-09-14 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | NRDE2 negatively regulates exosome functions by inhibiting MTR4 recruitment and exosome interaction.
Genes Dev., 33, 2019
|
|
6U0S
 
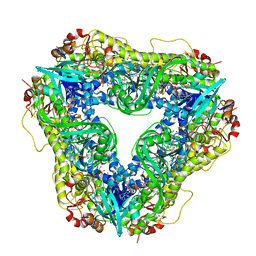 | | Crystal structure of the flavin-dependent monooxygenase PieE in complex with FAD and substrate | | Descriptor: | 2,4-dichlorophenol 6-monooxygenase, 2-[(2E,5E,7E,9R,10R,11E)-10-hydroxy-3,7,9,11-tetramethyltrideca-2,5,7,11-tetraen-1-yl]-6-methoxy-3-methylpyridin-4-ol, CHLORIDE ION, ... | | Authors: | Shi, R, Manenda, M. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural analyses of the Group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J.Biol.Chem., 295, 2020
|
|
6U0P
 
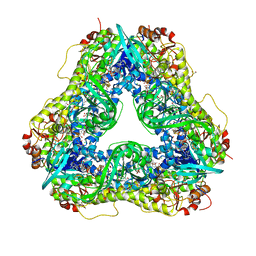 | | Crystal structure of PieE, the flavin-dependent monooxygenase involved in the biosynthesis of piericidin A1 | | Descriptor: | 2,4-dichlorophenol 6-monooxygenase, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Shi, R, Manenda, M, Picard, M.-E. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural analyses of the Group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J.Biol.Chem., 295, 2020
|
|
4ONA
 
 | |
6ILG
 
 | |
5T0Q
 
 | | Crystal structure of the Myc3 N-terminal domain [44-242] in complex with JAZ10 Jas domain [166-192] from arabidopsis | | Descriptor: | Protein TIFY 9, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J.S, He, S.Y, Xu, H.E, Melcher, K. | | Deposit date: | 2016-08-16 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6JYT
 
 | |
4RQW
 
 | | Crystal structure of Myc3 N-terminal JAZ-binding domain [44-238] from Arabidopsis | | Descriptor: | CALCIUM ION, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2014-11-05 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
6ILF
 
 | |
7MCF
 
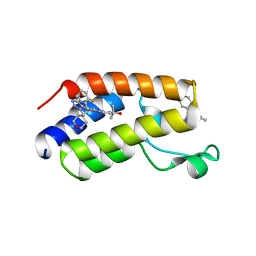 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH 2-{3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-6-fluoro-5-[(S)-(3-fluoropyridin-2-yl)(oxan-4-yl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol | | Descriptor: | 2-{3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-6-fluoro-5-[(S)-(3-fluoropyridin-2-yl)(oxan-4-yl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol, Bromodomain-containing protein 4 | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-02 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Development of BET inhibitors as potential treatments for cancer: A search for structural diversity.
Bioorg.Med.Chem.Lett., 44, 2021
|
|
4RRU
 
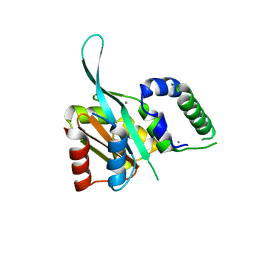 | | Myc3 N-terminal JAZ-binding domain[5-242] from arabidopsis | | Descriptor: | CALCIUM ION, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J.S, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2014-11-06 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
6ILE
 
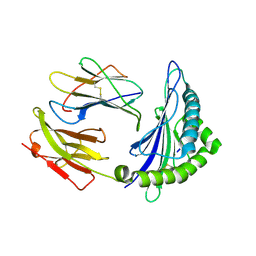 | | CRYSTAL STRUCTURE OF A MUTANT PTAL-N*01:01 FOR 2.9 ANGSTROM, 52M 53D 54L DELETED | | Descriptor: | Beta-2-microglobulin, HEV-1, MHC class I antigen | | Authors: | Qu, Z.H, Zhang, N.Z, Xia, C. | | Deposit date: | 2018-10-17 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and Peptidome of the Bat MHC Class I Molecule Reveal a Novel Mechanism Leading to High-Affinity Peptide Binding.
J Immunol., 202, 2019
|
|
6DBK
 
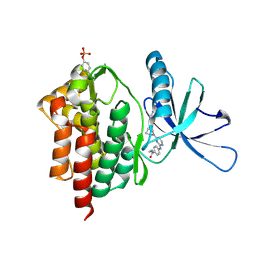 | | Tyk2 with compound 8 | | Descriptor: | 4-({4-[(1S,4S)-5-(cyanoacetyl)-2,5-diazabicyclo[2.2.1]heptan-2-yl]pyrimidin-2-yl}amino)-N-ethylbenzamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Vajdos, F.F. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-29 | | Last modified: | 2018-11-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dual Inhibition of TYK2 and JAK1 for the Treatment of Autoimmune Diseases: Discovery of (( S)-2,2-Difluorocyclopropyl)((1 R,5 S)-3-(2-((1-methyl-1 H-pyrazol-4-yl)amino)pyrimidin-4-yl)-3,8-diazabicyclo[3.2.1]octan-8-yl)methanone (PF-06700841).
J. Med. Chem., 61, 2018
|
|
5T0F
 
 | | Crystal structure of the Myc3 N-terminal domain [44-242] in complex with JAZ10 CMID domain [16-58] from arabidopsis | | Descriptor: | Protein TIFY 9, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J.S, He, S.Y, Xu, H.E, Melcher, K. | | Deposit date: | 2016-08-16 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4RS9
 
 | | Structure of Myc3 N-terminal JAZ-binding domain [44-238] in complex with Jas motif of JAZ9 | | Descriptor: | Protein TIFY 7, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J.S, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2014-11-07 | | Release date: | 2015-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
6DBM
 
 | | Tyk2 with compound 23 | | Descriptor: | Non-receptor tyrosine-protein kinase TYK2, [(1S)-2,2-difluorocyclopropyl][(1R,5S)-3-{2-[(1-methyl-1H-pyrazol-4-yl)amino]pyrimidin-4-yl}-3,8-diazabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Vajdos, F.F. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-29 | | Last modified: | 2018-11-07 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | Dual Inhibition of TYK2 and JAK1 for the Treatment of Autoimmune Diseases: Discovery of (( S)-2,2-Difluorocyclopropyl)((1 R,5 S)-3-(2-((1-methyl-1 H-pyrazol-4-yl)amino)pyrimidin-4-yl)-3,8-diazabicyclo[3.2.1]octan-8-yl)methanone (PF-06700841).
J. Med. Chem., 61, 2018
|
|
7CSD
 
 | | AtPrR1 with NADP+ and (+)lariciresinol | | Descriptor: | 4-[[(3R,4R,5S)-4-(hydroxymethyl)-5-(3-methoxy-4-oxidanyl-phenyl)oxolan-3-yl]methyl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79709971 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
