5JYK
 
 | | Deg9 crystal under 289K | | Descriptor: | GLYCEROL, Protease Do-like 9 | | Authors: | Ouyang, M, Zhang, L.X. | | Deposit date: | 2016-05-14 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
5K24
 
 | |
5K25
 
 | |
9LYC
 
 | | Cryo-EM structure of GPR3-G protein-dimer complex | | Descriptor: | G-protein coupled receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Hua, T, Liu, Z.J, Li, X.T, Chang, H. | | Deposit date: | 2025-02-19 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of oligomerization-modulated activation and autoinhibition of orphan receptor GPR3.
Cell Rep, 44, 2025
|
|
1WPQ
 
 | | Ternary Complex Of Glycerol 3-phosphate Dehydrogenase 1 with NAD and dihydroxyactone | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Glycerol-3-phosphate dehydrogenase [NAD+], cytoplasmic, ... | | Authors: | Ou, X, Han, X, Rao, Z. | | Deposit date: | 2004-09-10 | | Release date: | 2006-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Human Glycerol 3-phosphate Dehydrogenase 1 (GPD1)
J.Mol.Biol., 357, 2006
|
|
1X0V
 
 | |
1X0X
 
 | |
6M1H
 
 | | CryoEM structure of human PAC1 receptor in complex with maxadilan | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
7POI
 
 | | Prodomain bound BMP10 crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bone morphogenetic protein 10, D(-)-TARTARIC ACID | | Authors: | Guo, J, Yu, M, Li, W. | | Deposit date: | 2021-09-09 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of BMPRII extracellular domain in binary and ternary receptor complexes with BMP10.
Nat Commun, 13, 2022
|
|
7POJ
 
 | | Prodomain bound BMP10 crystal form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bone morphogenetic protein 10, TETRAETHYLENE GLYCOL | | Authors: | Guo, J, Yu, M, Li, W. | | Deposit date: | 2021-09-09 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of BMPRII extracellular domain in binary and ternary receptor complexes with BMP10.
Nat Commun, 13, 2022
|
|
7PPB
 
 | |
7PPA
 
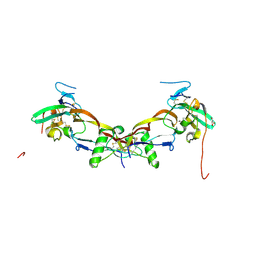 | | High resolution structure of bone morphogenetic protein receptor type II (BMPRII) extracellular domain in complex with BMP10 | | Descriptor: | Bone morphogenetic protein 10, Bone morphogenetic protein receptor type-2, GLYCEROL | | Authors: | Guo, J, Yu, M, Read, R.J, Li, W. | | Deposit date: | 2021-09-13 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of BMPRII extracellular domain in binary and ternary receptor complexes with BMP10.
Nat Commun, 13, 2022
|
|
6L8O
 
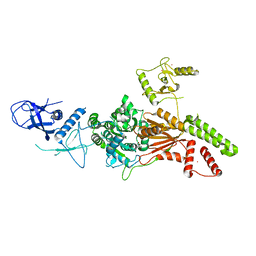 | | Crystal structure of the K. lactis Rad5 (Hg-derivative) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA repair protein RAD5, MERCURY (II) ION | | Authors: | Shen, M, Xiang, S. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
6L8N
 
 | | Crystal structure of the K. lactis Rad5 | | Descriptor: | DNA repair protein RAD5, ZINC ION | | Authors: | Shen, M, Xiang, S. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
6M1I
 
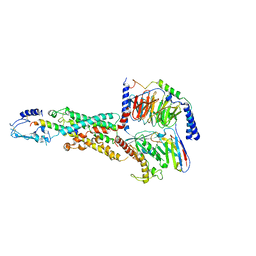 | | CryoEM structure of human PAC1 receptor in complex with PACAP38 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
9IJ1
 
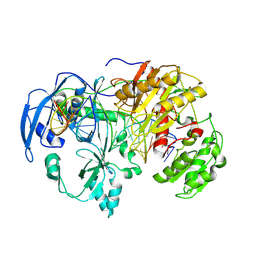 | | Cryo-EM Structure of MILI-piRNA-target (22-nt, bilobed) | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, RNA (26-MER), ... | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IIY
 
 | | Cryo-EM Structure of EfPiwi-piRNA-target (25-nt, bilobed) | | Descriptor: | Piwi, RNA (5'-R(P*CP*GP*UP*CP*UP*AP*UP*AP*CP*AP*AP*CP*CP*GP*AP*UP*CP*AP*GP*CP*U)-3'), RNA (5'-R(P*UP*AP*GP*CP*AP*GP*AP*UP*CP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*AP*CP*G)-3') | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IJ4
 
 | | Cryo-EM Structure of MILI(K852A)-piRNA-target (26-nt) | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, RNA (25-MER), ... | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
3E3R
 
 | |
7PPC
 
 | | Ternary signalling complex of BMP10 bound to ALK1 and BMPRII | | Descriptor: | Bone morphogenetic protein 10, Bone morphogenetic protein receptor type-2, Serine/threonine-protein kinase receptor R3 | | Authors: | Guo, J, Yu, M, Read, R.J, Li, W. | | Deposit date: | 2021-09-13 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structures of BMPRII extracellular domain in binary and ternary receptor complexes with BMP10.
Nat Commun, 13, 2022
|
|
9IJ5
 
 | | Cryo-EM Structure of MILI(K853A)-piRNA-target | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, RNA (25-MER), ... | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IJ3
 
 | | Cryo-EM Structure of MILI-piRNA-target (26-nt) | | Descriptor: | MANGANESE (II) ION, Piwi-like protein 2, RNA (25-MER), ... | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IJ0
 
 | | Cryo-EM Structure of MILI-piRNA-target (8-nt) | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, RNA (26-MER), ... | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IJ2
 
 | | Cryo-EM Structure of MILI-piRNA-target (22-nt, comma) | | Descriptor: | Piwi-like protein 2, RNA (5'-R(P*CP*AP*AP*GP*UP*UP*UP*CP*CP*AP*UP*GP*UP*UP*GP*AP*UP*GP*GP*UP*A)-3'), RNA (5'-R(P*UP*UP*AP*CP*CP*AP*UP*CP*AP*AP*CP*AP*UP*GP*GP*AP*AP*AP*CP*UP*UP*G)-3') | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IIZ
 
 | | Cryo-EM Structure of EfPiwi-piRNA-target (25-nt, comma) | | Descriptor: | Piwi, RNA (25-MER), RNA (5'-R(P*AP*GP*CP*CP*AP*AP*GP*UP*UP*UP*CP*CP*AP*UP*GP*UP*UP*GP*AP*UP*GP*GP*UP*A)-3') | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
