6JPQ
 
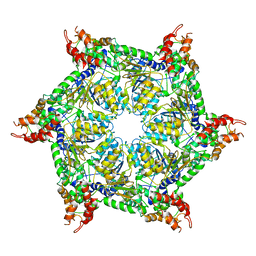 | | CryoEM structure of Abo1 hexamer - ADP complex | | Descriptor: | Uncharacterized AAA domain-containing protein C31G5.19 | | Authors: | Cho, C, Jang, J, Song, J.J. | | Deposit date: | 2019-03-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.44 Å) | | Cite: | Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone.
Nat Commun, 10, 2019
|
|
6JPU
 
 | | CryoEM structure of Abo1 hexamer - apo complex | | Descriptor: | Uncharacterized AAA domain-containing protein C31G5.19 | | Authors: | Cho, C, Jang, J, Song, J.J. | | Deposit date: | 2019-03-28 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone.
Nat Commun, 10, 2019
|
|
1D3U
 
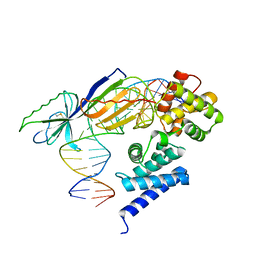 | | TATA-BINDING PROTEIN/TRANSCRIPTION FACTOR (II)B/BRE+TATA-BOX COMPLEX FROM PYROCOCCUS WOESEI | | Descriptor: | DNA 23-MER: BRE+TATA-BOX, DNA 24-MER: BRE+TATA-BOX, TATA-BINDING PROTEIN, ... | | Authors: | Littlefield, O, Korkhin, Y, Sigler, P.B. | | Deposit date: | 1999-10-01 | | Release date: | 1999-11-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis for the oriented assembly of a TBP/TFB/promoter complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1A2X
 
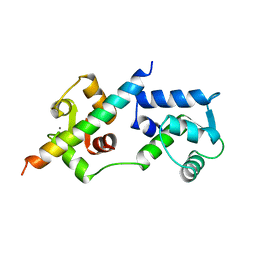 | | COMPLEX OF TROPONIN C WITH A 47 RESIDUE (1-47) FRAGMENT OF TROPONIN I | | Descriptor: | CALCIUM ION, TROPONIN C, TROPONIN I | | Authors: | Vassylyev, D.G, Takeda, S, Wakatsuki, S, Maeda, K, Maeda, Y. | | Deposit date: | 1998-01-13 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of troponin C in complex with troponin I fragment at 2.3-A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6JQ0
 
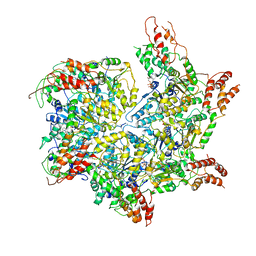 | | CryoEM structure of Abo1 Walker B (E372Q) mutant hexamer - ATP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Uncharacterized AAA domain-containing protein C31G5.19, ... | | Authors: | Cho, C, Jang, J, Song, J.J. | | Deposit date: | 2019-03-28 | | Release date: | 2019-12-25 | | Last modified: | 2020-01-01 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone.
Nat Commun, 10, 2019
|
|
5Z25
 
 | | Trimeric Alpha-Helix-Inserted Circular Permutant of Cytochrome c555 | | Descriptor: | Cytochrome c552, HEME C, TETRAETHYLENE GLYCOL | | Authors: | Oda, A, Nagao, S, Yamanaka, M, Ueda, I, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-12-28 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Construction of a Triangle-Shaped Trimer and a Tetrahedron Using an alpha-Helix-Inserted Circular Permutant of Cytochrome c555.
Chem Asian J, 13, 2018
|
|
1C0T
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH BM+21.1326 | | Descriptor: | (R)-(+)9B-(3-METHYL)PHENYL-2,3-DIHYDROTHIAZOLO[2,3-A]ISOINDOL-5(9BH)-ONE, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Ren, J, Esnouf, R.M, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-19 | | Release date: | 2000-07-19 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic analysis of the binding modes of thiazoloisoindolinone non-nucleoside inhibitors to HIV-1 reverse transcriptase and comparison with modeling studies.
J.Med.Chem., 42, 1999
|
|
2DEK
 
 | |
2DQX
 
 | | mutant beta-amylase (W55R) from soy bean | | Descriptor: | Beta-amylase | | Authors: | Ishikawa, K. | | Deposit date: | 2006-06-01 | | Release date: | 2007-05-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetic and structural analysis of enzyme sliding on a substrate: multiple attack in beta-amylase
Biochemistry, 46, 2007
|
|
1TBO
 
 | | NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, 30 STRUCTURES | | Descriptor: | PROTEIN KINASE C, GAMMA TYPE, ZINC ION | | Authors: | Xu, R.X, Pawelczyk, T, Xia, T, Brown, S.C. | | Deposit date: | 1997-04-15 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a protein kinase C-gamma phorbol-binding domain and study of protein-lipid micelle interactions.
Biochemistry, 36, 1997
|
|
5DGQ
 
 | |
1POZ
 
 | | SOLUTION STRUCTURE OF THE HYALURONAN BINDING DOMAIN OF HUMAN CD44 | | Descriptor: | CD44 antigen | | Authors: | Teriete, P, Banerji, S, Blundell, C.D, Kahmann, J.D, Pickford, A.R, Wright, A.J, Campbell, I.D, Jackson, D.G, Day, A.J. | | Deposit date: | 2003-06-16 | | Release date: | 2004-03-16 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the Regulatory Hyaluronan Binding Domain in the Inflammatory Leukocyte Homing Receptor CD44.
Mol.Cell, 13, 2004
|
|
1O7C
 
 | | Solution structure of the human TSG-6 Link module in the presence of a hyaluronan octasaccharide | | Descriptor: | TUMOR NECROSIS FACTOR-INDUCIBLE PROTEIN TSG-6 | | Authors: | Blundell, C.D, Teriete, P, Kahmann, J.D, Pickford, A.R, Campbell, I.D, Day, A.J. | | Deposit date: | 2002-10-29 | | Release date: | 2003-10-23 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | The link module from ovulation- and inflammation-associated protein TSG-6 changes conformation on hyaluronan binding.
J. Biol. Chem., 278, 2003
|
|
1O7B
 
 | | Refined solution structure of the human TSG-6 Link module | | Descriptor: | TUMOR NECROSIS FACTOR-INDUCIBLE PROTEIN TSG-6 | | Authors: | Blundell, C.D, Teriete, P, Kahmann, J.D, Pickford, A.R, Campbell, I.D, Day, A.J. | | Deposit date: | 2002-10-29 | | Release date: | 2003-10-23 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | The link module from ovulation- and inflammation-associated protein TSG-6 changes conformation on hyaluronan binding.
J. Biol. Chem., 278, 2003
|
|
5DGR
 
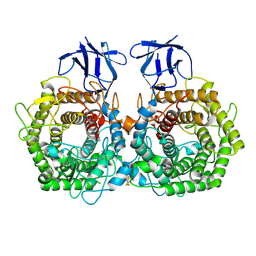 | | Crystal structure of GH9 exo-beta-D-glucosaminidase PBPRA0520, glucosamine complex | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, Putative endoglucanase-related protein, SODIUM ION | | Authors: | Suzuki, K, Honda, Y, Fushinobu, S. | | Deposit date: | 2015-08-28 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of an inverting glycoside hydrolase family 9 exo-beta-D-glucosaminidase and the design of glycosynthase.
Biochem.J., 473, 2016
|
|
1RMS
 
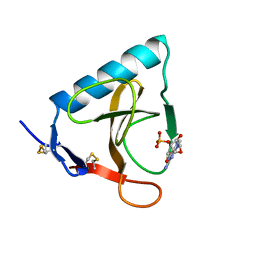 | | CRYSTAL STRUCTURES OF RIBONUCLEASE MS COMPLEXED WITH 3'-GUANYLIC ACID A GP*C ANALOGUE, 2'-DEOXY-2'-FLUOROGUANYLYL-3',5'-CYTIDINE | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE MS | | Authors: | Nonaka, T, Mitsui, Y, Nakamura, K.T. | | Deposit date: | 1991-12-02 | | Release date: | 1992-07-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ribonuclease Ms (as a ribonuclease T1 homologue) complexed with a guanylyl-3',5'-cytidine analogue.
Biochemistry, 32, 1993
|
|
8PAZ
 
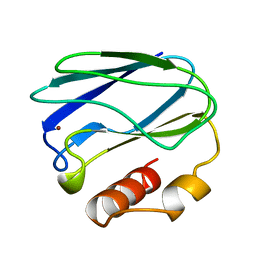 | | OXIDIZED NATIVE PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-24 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
1TBN
 
 | | NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN KINASE C, GAMMA TYPE, ZINC ION | | Authors: | Xu, R.X, Pawelczyk, T, Xia, T, Brown, S.C. | | Deposit date: | 1997-04-15 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a protein kinase C-gamma phorbol-binding domain and study of protein-lipid micelle interactions.
Biochemistry, 36, 1997
|
|
3WPF
 
 | | Crystal structure of mouse TLR9 (unliganded form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Toll-like receptor 9 | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.959 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3WPG
 
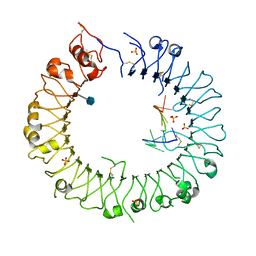 | |
3WPI
 
 | | Crystal structure of mouse TLR9 in complex with inhibitory DNA_super | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*CP*CP*TP*CP*AP*AP*TP*AP*GP*GP*GP*TP*GP*AP*GP*GP*GP*G)-3'), Toll-like receptor 9 | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3WPH
 
 | |
2Z7R
 
 | |
2Z7Q
 
 | |
2Z7S
 
 | | Crystal Structure of the N-terminal Kinase Domain of Human RSK1 bound to Purvalnol A | | Descriptor: | 2-({6-[(3-CHLOROPHENYL)AMINO]-9-ISOPROPYL-9H-PURIN-2-YL}AMINO)-3-METHYLBUTAN-1-OL, Ribosomal protein S6 kinase alpha-1 | | Authors: | Ikuta, M, Munshi, S.K. | | Deposit date: | 2007-08-28 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the N-terminal kinase domain of human RSK1 bound to three different ligands: Implications for the design of RSK1 specific inhibitors.
Protein Sci., 16, 2007
|
|
