5EDX
 
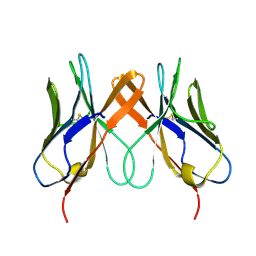 | | Crystal structure of swine CD8aa homodimer | | Descriptor: | CD8 alpha antigen | | Authors: | Liu, Y.J, Qi, J.X, Zhang, N.Z, Xia, C. | | Deposit date: | 2015-10-22 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The structural basis of chicken, swine and bovine CD8 alpha alpha dimers provides insight into the co-evolution with MHC I in endotherm species.
Sci Rep, 6, 2016
|
|
2OD1
 
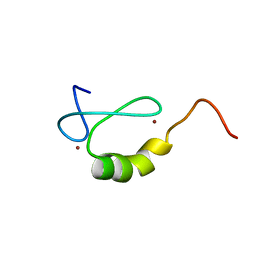 | | Solution structure of the MYND domain from human AML1-ETO | | Descriptor: | Protein CBFA2T1, ZINC ION | | Authors: | Liu, Y.Z, Chen, W, Gaudet, J, Cheney, M.D, Roudaia, L, Cierpicki, T, Klet, R.C, Hartman, K, Laue, T.M, Speck, N.A, Bushweller, J.H. | | Deposit date: | 2006-12-21 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO's activity.
Cancer Cell, 11, 2007
|
|
7KDC
 
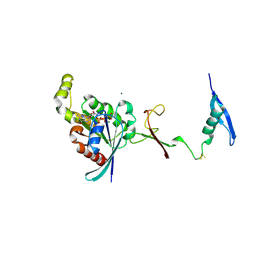 | | The complex between RhoD and the Plexin B2 RBD | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Plexin-B2, ... | | Authors: | Kuo, Y, Wang, Y, Zhang, x. | | Deposit date: | 2020-10-08 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A putative structural mechanism underlying the antithetic effect of homologous RND1 and RhoD GTPases in mammalian plexin regulation.
Elife, 10, 2021
|
|
8YZD
 
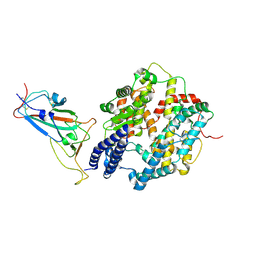 | | Structure of JN.1 RBD protein in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,fusion protein | | Authors: | Wang, Y.J, Zhang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
8YZE
 
 | | The JN.1 spike protein (S) in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,Expression Tag | | Authors: | Wang, Y.J, Zhang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
8YZC
 
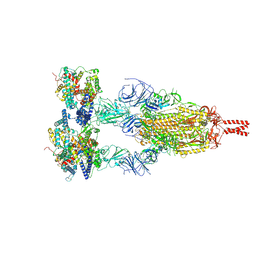 | | Structure of BA.2.86 spike protein in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,Expression Tag | | Authors: | Wang, Y.J, Zang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
8YZB
 
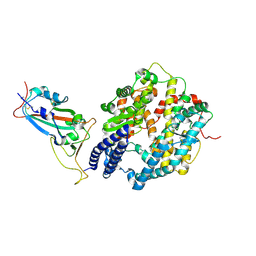 | | BA.2.86 RBD protein in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,fusion protein | | Authors: | Wang, Y.J, Zhang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
5CNQ
 
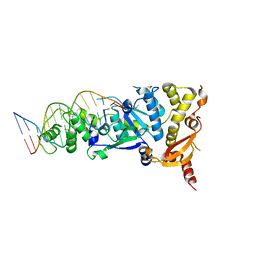 | | Crystal structure of the Holliday junction-resolving enzyme GEN1 (WT) in complex with product DNA, Mg2+ and Mn2+ ions | | Descriptor: | DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), MANGANESE (II) ION, Nuclease-like protein, ... | | Authors: | Liu, Y.J, Freeman, A.D.J, Declais, A.C, Wilson, T.J, Gartner, A, Lilley, D.M.J. | | Deposit date: | 2015-07-17 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Crystal Structure of a Eukaryotic GEN1 Resolving Enzyme Bound to DNA.
Cell Rep, 13, 2015
|
|
5CO8
 
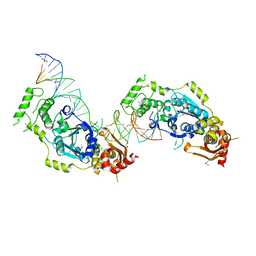 | | Crystal structure of the Holliday junction-resolving enzyme GEN1 (WT) in complex with product DNA and Mg2+ ion | | Descriptor: | DNA (31-MER), DNA (5'-D(*AP*GP*AP*CP*TP*GP*CP*AP*GP*TP*TP*GP*AP*GP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*A)-3'), ... | | Authors: | Liu, Y.J, Freeman, A.D.J, Declais, A.C, Wilson, T.J, Gartner, A, Lilley, D.M.J. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a Eukaryotic GEN1 Resolving Enzyme Bound to DNA.
Cell Rep, 13, 2015
|
|
6XZ8
 
 | | Structure of aldosterone synthase (CYP11B2) in complex with N-[(1R)-1-[5-(6-chloro-1,1-dimethyl-3-oxo-isoindolin-2-yl)-3-pyridyl]ethyl]methanesulfonamide | | Descriptor: | Cytochrome P450 11B2, mitochondrial, HEME C, ... | | Authors: | Kuglstatter, A, Joseph, C, Benz, J. | | Deposit date: | 2020-02-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of 3-Pyridyl Isoindolin-1-one Derivatives as Potent, Selective, and Orally Active Aldosterone Synthase (CYP11B2) Inhibitors.
J.Med.Chem., 63, 2020
|
|
5UXZ
 
 | |
1HSJ
 
 | | SARR MBP FUSION STRUCTURE | | Descriptor: | FUSION PROTEIN CONSISTING OF STAPHYLOCOCCUS ACCESSORY REGULATOR PROTEIN R AND MALTOSE BINDING PROTEIN, alpha-D-glucopyranose | | Authors: | Zhang, G. | | Deposit date: | 2000-12-26 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the SarR protein from Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6XZ9
 
 | | Structure of aldosterone synthase (CYP11B2) in complex with 5-chloro-3,3-dimethyl-2-[5-[1-(1-methylpyrazole-4-carbonyl)azetidin-3-yl]oxy-3-pyridyl]isoindolin-1-one | | Descriptor: | 5-chloranyl-3,3-dimethyl-2-[5-[1-(1-methylpyrazol-4-yl)carbonylazetidin-3-yl]oxypyridin-3-yl]isoindol-1-one, Cytochrome P450 11B2, mitochondrial, ... | | Authors: | Kuglstatter, A, Joseph, C, Benz, J. | | Deposit date: | 2020-02-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Discovery of 3-Pyridyl Isoindolin-1-one Derivatives as Potent, Selective, and Orally Active Aldosterone Synthase (CYP11B2) Inhibitors.
J.Med.Chem., 63, 2020
|
|
5UY1
 
 | | X-ray crystal structure of apo Halotag | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Dunham, N.P, Boal, A.K. | | Deposit date: | 2017-02-23 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Cation-pi Interaction Enables a Halo-Tag Fluorogenic Probe for Fast No-Wash Live Cell Imaging and Gel-Free Protein Quantification.
Biochemistry, 56, 2017
|
|
4FGX
 
 | | Crystal structure of bace1 with novel inhibitor | | Descriptor: | Beta-secretase 1, DI(HYDROXYETHYL)ETHER, Inhibitor (2R,5S,8S,12S,13S,16S,19S,22S)-16-(3-amino-3-oxopropyl)-2,13-dibenzyl-12,22-dihydroxy-8-isobutyl-19-isopropyl-3,5,17-trimethyl-4,7,10,15,18,21-hexaoxo-3,6,9,14,17,20-hexaazatricosan-1-oic acid, ... | | Authors: | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | Deposit date: | 2012-06-05 | | Release date: | 2013-01-16 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
6AGS
 
 | |
8K3Z
 
 | | Cryo-EM structure of CXCR4 in complex with CXCL12 | | Descriptor: | C-X-C chemokine receptor type 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Y.Z, Liu, A.J, Liao, Q.W, Ye, R.D. | | Deposit date: | 2023-07-17 | | Release date: | 2024-07-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-EM structure of monomeric CXCL12-bound CXCR4 in the active state.
Cell Rep, 43, 2024
|
|
4OU1
 
 | | Crystal structure of a computationally designed retro-aldolase covalently bound to folding probe 1 [(6-methoxynaphthalen-2-yl)(oxiran-2-yl)methanol] | | Descriptor: | (1S,2S)-1-(6-methoxynaphthalen-2-yl)propane-1,2-diol, BENZOIC ACID, PHOSPHATE ION, ... | | Authors: | Bhabha, G, Zhang, X, Ekiert, D.C. | | Deposit date: | 2014-02-14 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small molecule probes to quantify the functional fraction of a specific protein in a cell with minimal folding equilibrium shifts.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2ODD
 
 | | Solution structure of the MYND domain from AML1-ETO complexed with SMRT, a corepressor | | Descriptor: | Protein CBFA2T1, SMRT, ZINC ION | | Authors: | Liu, Y.Z, Chen, W, Gaudet, J, Cheney, M.D, Roudaia, L, Cierpicki, T, Klet, R.C, Hartman, K, Laue, T.M, Speck, N.A, Bushweller, J.H. | | Deposit date: | 2006-12-22 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO's activity.
Cancer Cell, 11, 2007
|
|
7VWV
 
 | |
6AZR
 
 | | Crystal structure of the T264A HK853cp-BeF3-RR468 complex | | Descriptor: | Chemotaxis regulator-transmits chemoreceptor signals to flagelllar motor components CheY, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Rose, J, Zhou, P. | | Deposit date: | 2017-09-11 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.628 Å) | | Cite: | A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases.
Nat Commun, 8, 2017
|
|
4DVF
 
 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2S)-1-[(2R,5S,8S,12S,13S)-2,13-DIBENZYL-12-HYDROXY-3,5-DIMETHYL-8-(2-METHYLPROPYL)-15-(3-[(METHYLSULFONYL)AMINO]-5-{[(1R)-1-PHENYLETHYL]CARBAMOYL}PHENYL)-4,7,10,15-TETRAOXO-3,6,9,14-TETRAAZAPENTADECAN-1-OYL]PYRROLIDINE-2-CARBOXYLATE | | Authors: | Xu, Y.C, Chen, W.Y, Li, L, Chen, T.T. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
4DV9
 
 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2S)-1-[(2R,5S,8S,12S,13S,16S,19S,22S)-16-(3-AMINO-3-OXOPROPYL)-2,13-DIBENZYL-12,22-DIHYDROXY-3,5,17-TRIMETHYL-8-(2-METHYLPROPYL)-4,7,10,15,18,21-HEXAOXO-19-(PROPAN-2-YL)-3,6,9,14,17,20-HEXAAZATRICOSAN-1-OYL]PYRROLIDINE-2-CARBOXYLATE (NON-PREFERRED NAME), SULFATE ION | | Authors: | Xu, Y.C, Chen, W.Y, Li, L, Chen, T.T. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (2.076 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
7DV8
 
 | | The crystal structure of rice immune receptor RGA5-HMA2. | | Descriptor: | Disease resistance protein RGA5 | | Authors: | Zhang, X, Liu, J.F. | | Deposit date: | 2021-01-12 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | A designer rice NLR immune receptor confers resistance to the rice blast fungus carrying noncorresponding avirulence effectors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6LCG
 
 | | Structure of D-carbamoylase mutant from Nitratireductor indicus | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-carbamoyl-D-amino-acid hydrolase | | Authors: | Liu, Y.F, Ni, Y, Xu, G.C, Dai, W. | | Deposit date: | 2019-11-18 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Guided Engineering of D-Carbamoylase Reveals a Key Loop at Substrate Entrance Tunnel
Acs Catalysis, 10, 2020
|
|
