3OMS
 
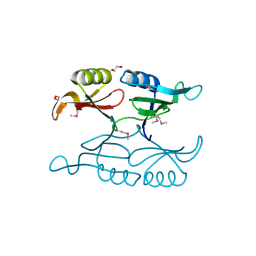 | | Putative 3-demethylubiquinone-9 3-methyltransferase, PhnB protein, from Bacillus cereus. | | Descriptor: | 1,2-ETHANEDIOL, PhnB protein | | Authors: | Osipiuk, J, Li, H, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-08 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystal structure of putative 3-demethylubiquinone-9
3-methyltransferase, PhnB protein, from Bacillus cereus.
To be Published
|
|
3ON3
 
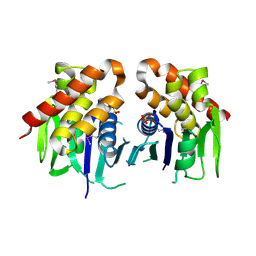 | | The crystal structure of keto/oxoacid ferredoxin oxidoreductase, gamma subunit from Geobacter sulfurreducens PCA | | Descriptor: | Keto/oxoacid ferredoxin oxidoreductase, gamma subunit, SULFATE ION | | Authors: | Tan, K, Zhang, R, Hatzos, C, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | The crystal structure of keto/oxoacid ferredoxin oxidoreductase, gamma subunit from Geobacter sulfurreducens PCA
To be Published
|
|
4QVS
 
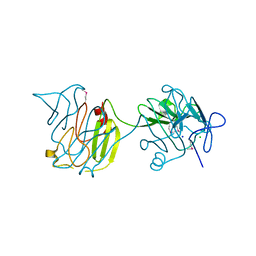 | | 2.1 Angstrom resolution crystal structure of S-layer domain-containing protein (residues 221-444) from Clostridium thermocellum ATCC 27405 | | Descriptor: | CHLORIDE ION, S-layer domain-containing protein, SODIUM ION | | Authors: | Halavaty, A.S, Wawrzak, Z, Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-15 | | Release date: | 2014-07-30 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom resolution crystal structure of S-layer domain-containing protein (residues 221-444) from Clostridium thermocellum ATCC 27405
To be Published
|
|
3ON4
 
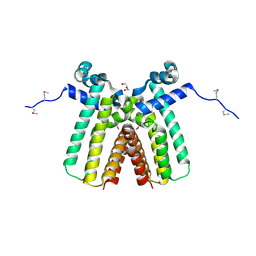 | | Crystal structure of TetR transcriptional regulator from Legionella pneumophila | | Descriptor: | DI(HYDROXYETHYL)ETHER, SODIUM ION, Transcriptional regulator, ... | | Authors: | Michalska, K, Li, H, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of TetR transcriptional regulator from Legionella pneumophila
To be Published
|
|
3MCZ
 
 | |
3M1G
 
 | | The structure of a putative glutathione S-transferase from Corynebacterium glutamicum | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Putative glutathione S-transferase | | Authors: | Cuff, M.E, Marshall, N, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-04 | | Release date: | 2010-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of a putative glutathione S-transferase from Corynebacterium glutamicum
TO BE PUBLISHED
|
|
1JI0
 
 | | Crystal Structure Analysis of the ABC transporter from Thermotoga maritima | | Descriptor: | ABC transporter, ADENOSINE-5'-TRIPHOSPHATE | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Beasley, S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-06-28 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A Crystal Structure of ABC Transporter from Thermotoga maritima
TO BE PUBLISHED
|
|
3MR0
 
 | | Crystal Structure of Sensory Box Histidine Kinase/Response Regulator from Burkholderia thailandensis E264 | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Tesar, C, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.493 Å) | | Cite: | Crystal Structure of Sensory Box Histidine Kinase/Response Regulator from Burkholderia thailandensis E264
To be Published
|
|
3MAH
 
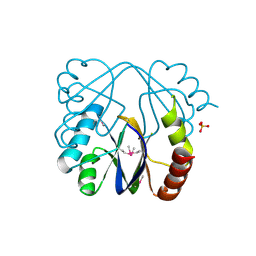 | | A putative c-terminal regulatory domain of aspartate kinase from porphyromonas gingivalis w83. | | Descriptor: | Aspartokinase, SULFATE ION | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Moy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A Putative C-Terminal Regulatory Domain of Aspartate Kinase from Porphyromonas Gingivalis W83.
To be Published
|
|
3NJN
 
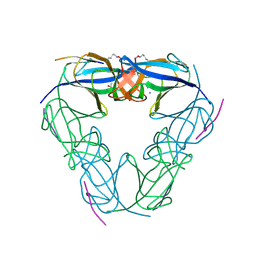 | | Q118A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
1MKI
 
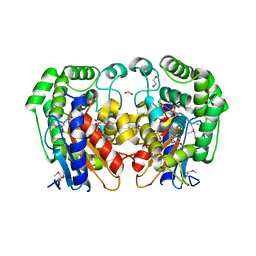 | | Crystal Structure of Bacillus Subtilis Probable Glutaminase, APC1040 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Probable Glutaminase ybgJ | | Authors: | Kim, Y, Dementieva, I, Vinokour, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-29 | | Release date: | 2003-06-03 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
3PEB
 
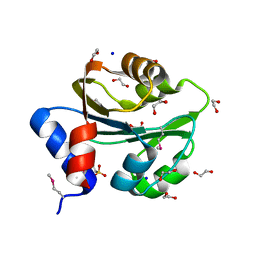 | | The Structure of a Creatine_N Superfamily domain of a dipeptidase from Streptococcus thermophilus. | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Dipeptidase, ... | | Authors: | Cuff, M.E, Mack, J.C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-10-25 | | Release date: | 2010-11-03 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Structure of a Creatine_N Superfamily domain of a dipeptidase from Streptococcus thermophilus.
TO BE PUBLISHED
|
|
1Q8B
 
 | |
3NKL
 
 | | Crystal Structure of UDP-D-Quinovosamine 4-Dehydrogenase from Vibrio fischeri | | Descriptor: | ACETIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Mack, J, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-20 | | Release date: | 2010-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of UDP-D-Quinovosamine 4-Dehydrogenase from Vibrio fischeri
To be Published, 2010
|
|
3MTJ
 
 | | The Crystal Structure of a Homoserine Dehydrogenase from Thiobacillus denitrificans to 2.15A | | Descriptor: | Homoserine dehydrogenase, SULFATE ION | | Authors: | Stein, A.J, Cui, H, Chin, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-30 | | Release date: | 2010-05-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of a Homoserine Dehydrogenase from Thiobacillus denitrificans to 2.15A
To be Published
|
|
3MWB
 
 | | The Crystal Structure of Prephenate dehydratase in complex with L-Phe from Arthrobacter aurescens to 2.0A | | Descriptor: | MAGNESIUM ION, PHENYLALANINE, Prephenate dehydratase | | Authors: | Stein, A.J, Chhor, G, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-05 | | Release date: | 2010-06-30 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Prephenate dehydratase in complex with L-Phe from Arthrobacter aurescens to 2.0A
To be Published
|
|
3MUX
 
 | | The Crystal Structure of a putative 4-hydroxy-2-oxoglutarate aldolase from Bacillus anthracis to 1.45A | | Descriptor: | CHLORIDE ION, SODIUM ION, putative 4-hydroxy-2-oxoglutarate aldolase | | Authors: | Stein, A.J, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-03 | | Release date: | 2010-05-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of a putative 4-hydroxy-2-oxoglutarate aldolase from Bacillus anthracis to 1.45A
To be Published
|
|
3MY7
 
 | | The Crystal Structure of the ACDH domain of an Alcohol Dehydrogenase from Vibrio parahaemolyticus to 2.25A | | Descriptor: | Alcohol dehydrogenase/acetaldehyde dehydrogenase, CHLORIDE ION | | Authors: | Stein, A.J, Weger, A, Volkart, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-10 | | Release date: | 2010-06-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the ACDH domain of an Alcohol Dehydrogenase from Vibrio parahaemolyticus to 2.25A
To be Published
|
|
3N73
 
 | | Crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase from Bacillus cereus | | Descriptor: | CHLORIDE ION, Putative 4-hydroxy-2-oxoglutarate aldolase | | Authors: | Cabello, R, Chruszcz, M, Xu, X, Zimmerman, M.D, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-26 | | Release date: | 2010-06-09 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase from Bacillus cereus
To be Published
|
|
3PU9
 
 | | Crystal structure of serine/threonine phosphatase Sphaerobacter thermophilus DSM 20745 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Protein serine/threonine phosphatase | | Authors: | Nocek, B, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-22 | | Last modified: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of serine/threonine phosphatase Sphaerobacter thermophilus DSM 20745
TO BE PUBLISHED
|
|
3PNN
 
 | |
3NJH
 
 | | D37A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis. | | Descriptor: | CALCIUM ION, GLYCEROL, Peptidase, ... | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
3N70
 
 | | The Crystal Structure of the P-loop NTPase domain of the Sigma-54 transport activator from E. coli to 2.8A | | Descriptor: | SULFATE ION, Transport activator | | Authors: | Stein, A.J, Mulligan, R, Volkart, L, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-26 | | Release date: | 2010-07-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the P-loop NTPase domain of the Sigma-54 transport activator from E. coli to 2.8A
To be Published
|
|
4F06
 
 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris HaA2 RPB_2270 in complex with P-HYDROXYBENZOIC ACID | | Descriptor: | Extracellular ligand-binding receptor, GLYCEROL, P-HYDROXYBENZOIC ACID, ... | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-03 | | Release date: | 2012-08-15 | | Last modified: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of transport proteins for aromatic compounds derived from lignin: benzoate derivative binding proteins.
J.Mol.Biol., 423, 2012
|
|
4EYK
 
 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris BisB5 in complex with 3,4-dihydroxy benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 3,4-DIHYDROXYBENZOIC ACID, Twin-arginine translocation pathway signal | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of transport proteins for aromatic compounds derived from lignin: benzoate derivative binding proteins.
J.Mol.Biol., 423, 2012
|
|
