9K36
 
 | | Human RNA Polymerase III de novo transcribing complex 12 (TC12) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (54-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action.
Nature, 2025
|
|
9LXO
 
 | | Human RNA Polymerase III de novo transcribing complex 6 (TC6)(without 3'dATP) | | Descriptor: | DNA (127-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2025-02-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action.
Nature, 2025
|
|
9K39
 
 | | Human RNA Polymerase III de novo transcribing complex 8 (TC8) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (50-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action.
Nature, 2025
|
|
9K2G
 
 | | Human RNA Polymerase III de novo transcribing complex 13 (TC13) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (104-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-17 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action.
Nature, 2025
|
|
9K3U
 
 | | Human RNA Polymerase III de novo transcribing complex 5 (TC5) | | Descriptor: | DNA (97-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action.
Nature, 2025
|
|
9K38
 
 | | Human RNA Polymerase III de novo transcribing complex 6 (TC6) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (51-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action.
Nature, 2025
|
|
9K3B
 
 | | Human RNA Polymerase III de novo transcribing complex 10 overall (TC10-overall) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (102-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action.
Nature, 2025
|
|
4Y93
 
 | | Crystal structure of the PH-TH-kinase construct of Bruton's tyrosine kinase (Btk) | | Descriptor: | 4-tert-butyl-N-[2-methyl-3-(4-methyl-6-{[4-(morpholin-4-ylcarbonyl)phenyl]amino}-5-oxo-4,5-dihydropyrazin-2-yl)phenyl]benzamide, CALCIUM ION, Non-specific protein-tyrosine kinase,Non-specific protein-tyrosine kinase, ... | | Authors: | Wang, Q, Kuriyan, J. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Autoinhibition of Bruton's tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate.
Elife, 4, 2015
|
|
8T3P
 
 | |
7Y9M
 
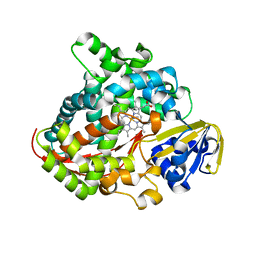 | | Crystal structure of P450 BM3-2F from Bacillus megaterium | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, Q, Zhang, L.L, Liu, W.D, Huang, J.-W, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-25 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of P450 BM3-2F from Bacillus megaterium
to be published
|
|
7Y9L
 
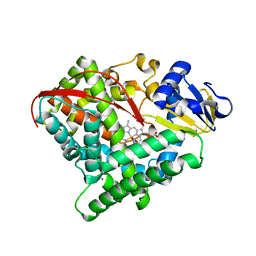 | | Crystal structure of P450 BM3-2F from Bacillus megaterium in complex with 2-Hydroxy-5-Nitrobenzonitrile | | Descriptor: | 5-nitro-2-oxidanyl-benzenecarbonitrile, Bifunctional cytochrome P450/NADPH--P450 reductase, NICKEL (II) ION, ... | | Authors: | Wang, Q, Zhang, L.L, Liu, W.D, Huang, J.-W, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-25 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of P450 BM3-2F from Bacillus megaterium in complex with 2-Hydroxy-5-Nitrobenzonitrile
to be published
|
|
3SVR
 
 | |
3SVS
 
 | |
3SVN
 
 | |
3SVU
 
 | |
3SVO
 
 | |
5XYB
 
 | |
5Y24
 
 | | Crystal structure of AimR from Bacillus phage SPbeta in complex with its signalling peptide | | Descriptor: | AimR transcriptional regulator, BROMIDE ION, GLY-MET-PRO-ARG-GLY-ALA | | Authors: | Wang, Q, Guan, Z.Y, Zou, T.T, Yin, P. | | Deposit date: | 2017-07-24 | | Release date: | 2018-09-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structural basis of the arbitrium peptide-AimR communication system in the phage lysis-lysogeny decision.
Nat Microbiol, 3, 2018
|
|
6AEE
 
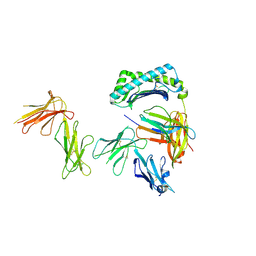 | | Crystal structure of the four Ig-like domains of LILRB1 complexed with HLA-G | | Descriptor: | 9 Mer Peptide (RL9) From Histone H2A.x, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Wang, Q, Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2018-08-04 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Structures of the four Ig-like domain LILRB2 and the four-domain LILRB1 and HLA-G1 complex.
Cell. Mol. Immunol., 2019
|
|
6AED
 
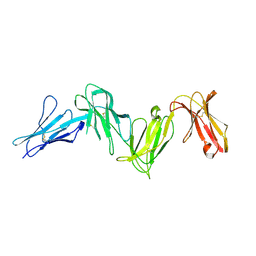 | |
2I0N
 
 | | Structure of Dictyostelium discoideum Myosin VII SH3 domain with adjacent proline rich region | | Descriptor: | Class VII unconventional myosin | | Authors: | Wang, Q, Deloia, M.A, Kang, Y, Litchke, C, Titus, M.A, Walters, K.J. | | Deposit date: | 2006-08-10 | | Release date: | 2007-01-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The SH3 domain of a M7 interacts with its C-terminal proline-rich region.
Protein Sci., 16, 2007
|
|
9IMQ
 
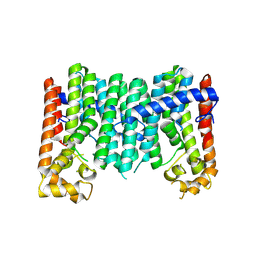 | | Crystal structure of a C45 isoprenyl diphosphate synthase, Rv0562 from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, Nonaprenyl diphosphate synthase | | Authors: | Wang, Q, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-07-04 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural insight of a bi-functional isoprenyl diphosphate synthase Rv0562 from Mycobacterium tuberculosis.
Int.J.Biol.Macromol., 2025
|
|
9IMR
 
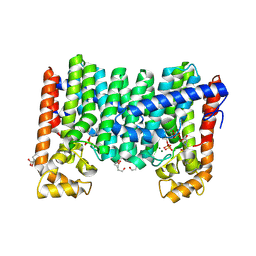 | | Crystal structure of geranylgeranyl pyrophosphate synthase Rv0562 from Mycobacterium tuberculosis in complex with IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, BICINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, Q, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-07-04 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insight of a bi-functional isoprenyl diphosphate synthase Rv0562 from Mycobacterium tuberculosis.
Int.J.Biol.Macromol., 2025
|
|
9IMS
 
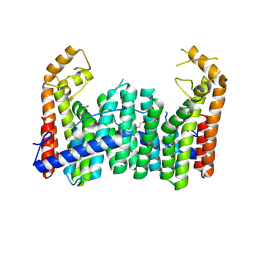 | |
8W5J
 
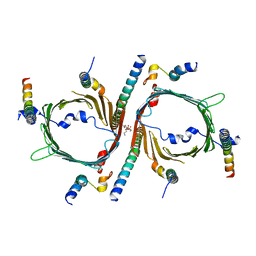 | | Cryo-EM structure of the yeast TOM core complex (from TOM-TIM23 complex) | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | Deposit date: | 2023-08-27 | | Release date: | 2024-02-28 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
