4REY
 
 | | Crystal Structure of the GRASP65-GM130 C-terminal peptide complex | | Descriptor: | Golgi reassembly-stacking protein 1, Golgin subfamily A member 2, SULFATE ION | | Authors: | Shi, N, Hu, F, Li, B. | | Deposit date: | 2014-09-24 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis for the Interaction between the Golgi Reassembly-stacking Protein GRASP65 and the Golgi Matrix Protein GM130.
J.Biol.Chem., 290, 2015
|
|
8ZYM
 
 | | Complex structure of 60 Fab bound to DS2 prefusion F trimer | | Descriptor: | Fusion glycoprotein F0,Expression tag, antibody light chain, antidody heavy chain | | Authors: | Wang, X, Ge, J, Guo, F. | | Deposit date: | 2024-06-18 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | DS2 designer pre-fusion F vaccine induces strong and protective antibody response against RSV infection.
Npj Vaccines, 9, 2024
|
|
5XTZ
 
 | | Crystal structure of GAS41 YEATS bound to H3K27ac peptide | | Descriptor: | ACETATE ION, THR-LYS-ALA-ALA-ARG-ALY-SER-ALA-PRO-ALA, YEATS domain-containing protein 4 | | Authors: | Li, H.T, Zhao, D. | | Deposit date: | 2017-06-21 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Gas41 links histone acetylation to H2A.Z deposition and maintenance of embryonic stem cell identity.
Cell Discov, 4, 2018
|
|
9J11
 
 | | Structure of mEos3.2 in the green fluorescent state | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Green to red photoconvertible GFP-like protein EosFP | | Authors: | Zheng, S.P, Shi, X.R. | | Deposit date: | 2024-08-03 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the fast maturation of pcStar, a photoconvertible fluorescent protein.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
9J0R
 
 | | Structure of pcStar in the green fluorescent state | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Green to red photoconvertible GFP-like protein EosFP | | Authors: | Zheng, S.P, Shi, X.R. | | Deposit date: | 2024-08-02 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis for the fast maturation of pcStar, a photoconvertible fluorescent protein.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
9J0Q
 
 | | Structure of mEos3.2 in the green fluorescent state | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Zheng, S.P, Shi, X.R. | | Deposit date: | 2024-08-02 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural basis for the fast maturation of pcStar, a photoconvertible fluorescent protein.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7F29
 
 | |
6FTR
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wiedorn, M.O, Oberthuer, D, Barty, A, Chapman, H.N. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.76000106 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
6GTH
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Wiedorn, M, Oberthuer, D, Werner, N, Schubert, R, White, T.A, Mancuso, A, Perbandt, M, Betzel, C, Barty, A, Chapman, H. | | Deposit date: | 2018-06-18 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
6GMA
 
 | |
8HPB
 
 | |
8HQB
 
 | | NMR Structure of OsCIE1-Ubox | | Descriptor: | U-box domain-containing protein 12 | | Authors: | Zhang, Y, Yu, C.Z, Lan, W.X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Release of a ubiquitin brake activates OsCERK1-triggered immunity in rice.
Nature, 629, 2024
|
|
7FJO
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with three T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7FJS
 
 | | Crystal structure of T6 Fab bound to theSARS-CoV-2 RBD of B.1.351 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7FJN
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with two T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7FJC
 
 | | Crystal structure of SARS-CoV-2 Beta RBD complexed with P36-5D2 Fab | | Descriptor: | P36-5D2 heavy chain, P36-5D2 light chain, Spike protein S1, ... | | Authors: | Zhang, L.Q, Wang, X.Q, Shan, S.S, Lan, J. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants.
Front Immunol, 12, 2021
|
|
8I4T
 
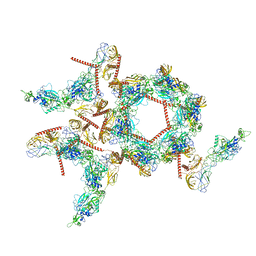 | | Structure of the asymmetric unit of SFTSV virion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein | | Authors: | Du, S, Peng, R, Qi, J, Li, C. | | Deposit date: | 2023-01-21 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-EM structure of severe fever with thrombocytopenia syndrome virus.
Nat Commun, 14, 2023
|
|
5XNV
 
 | | Crystal structure of YEATS2 YEATS bound to H3K27ac peptide | | Descriptor: | ALA-ALA-ARG-ALY-SER-ALA-PRO-ALA, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Li, H.T, Guan, H.P, Zhao, D. | | Deposit date: | 2017-05-24 | | Release date: | 2017-11-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | YEATS2 links histone acetylation to tumorigenesis of non-small cell lung cancer.
Nat Commun, 8, 2017
|
|
8HT7
 
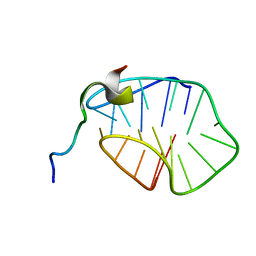 | | The N-terminal region of Cdc6 specifically recognizes human DNA G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*TP*GP*GP*G)-3'), GLN-ALA-GLN-ALA-THR-ILE-SER-PHE-PRO-LYS-ARG-LYS-LEU-SER-TRP | | Authors: | Liu, C, Zhu, G, Geng, Y, Xu, N. | | Deposit date: | 2022-12-20 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The N-terminal region of Cdc6 specifically recognizes human DNA G-quadruplex.
Int.J.Biol.Macromol., 260, 2024
|
|
5J9S
 
 | | ENL YEATS in complex with histone H3 acetylation at K27 | | Descriptor: | H3K27ac peptide from Histone H3.1, Protein ENL, SULFATE ION | | Authors: | Li, Y, Li, H. | | Deposit date: | 2016-04-11 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia
Nature, 543, 2017
|
|
5B73
 
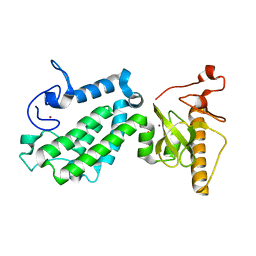 | | Crystal structure of human ZMYND8 PHD-Bromo-PWWP domain | | Descriptor: | Protein kinase C-binding protein 1, ZINC ION | | Authors: | Li, H, Li, Y, Zheng, X. | | Deposit date: | 2016-06-03 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ZMYND8 Reads the Dual Histone Mark H3K4me1-H3K14ac to Antagonize the Expression of Metastasis-Linked Genes
Mol.Cell, 63, 2016
|
|
6LSB
 
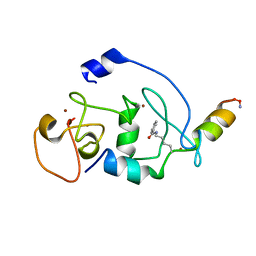 | |
6OIE
 
 | |
9B7A
 
 | | Crystal structure of peroxiredoxin 1 with RA | | Descriptor: | (2R)-3-(3,4-dihydroxyphenyl)-2-{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}propanoic acid, Peroxiredoxin-1 | | Authors: | Wu, Y, Xu, H, Luo, C. | | Deposit date: | 2024-03-27 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Inhibited peroxidase activity of peroxiredoxin 1 by palmitic acid exacerbates nonalcoholic steatohepatitis in male mice.
Nat Commun, 16, 2025
|
|
8HGA
 
 | |
