7BVE
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbC2-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, Ethambutol, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVG
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with di-arabinose. | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
6KQX
 
 | | Crystal structure of Yijc from B. subtilis in complex with UDP | | Descriptor: | URIDINE-5'-DIPHOSPHATE, Uncharacterized UDP-glucosyltransferase YjiC | | Authors: | Hu, Y.M, Dai, L.H, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural dissection of unnatural ginsenoside-biosynthetic UDP-glycosyltransferase Bs-YjiC from Bacillus subtilis for substrate promiscuity.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
6KQW
 
 | | Crystal structure of Yijc from B. subtilis | | Descriptor: | CITRIC ACID, Uncharacterized UDP-glucosyltransferase YjiC | | Authors: | Hu, Y.M, Dai, L.H, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural dissection of unnatural ginsenoside-biosynthetic UDP-glycosyltransferase Bs-YjiC from Bacillus subtilis for substrate promiscuity.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7PQY
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with FI-3A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, FI-3A Fab heavy chain, FI-3A Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
7PQZ
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with FI-3A and FD-11A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FD-11A Fab heavy chain, FD-11A Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
7PR0
 
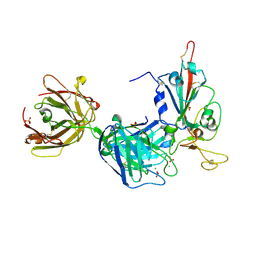 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with FD-5D Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, FD-5D Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
7Q0A
 
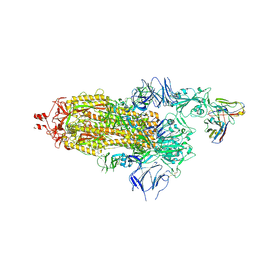 | | SARS-CoV-2 Spike ectodomain with Fab FI3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FI3A fab Light chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
6YSE
 
 | |
7WEE
 
 | |
7WEB
 
 | |
7WE9
 
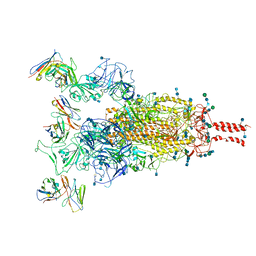 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7WLC
 
 | |
7WED
 
 | |
7WEA
 
 | |
7WEF
 
 | |
7WEC
 
 | |
7WE8
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv265 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 265, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7WE7
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv282 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 282, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7LV3
 
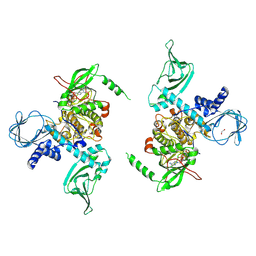 | | Crystal structure of human protein kinase G (PKG) R-C complex in inhibited state | | Descriptor: | 1,2-ETHANEDIOL, Isoform Beta of cGMP-dependent protein kinase 1, MANGANESE (II) ION, ... | | Authors: | Sharma, R, Lying, Q, Casteel, D, Kim, C. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | An auto-inhibited state of protein kinase G and implications for selective activation.
Elife, 11, 2022
|
|
5HFM
 
 | | Gp41-targeting HIV-1 fusion inhibitors with hook-like Ile-Asp-Leu tail | | Descriptor: | Envelope glycoprotein gp160,gp41 CHR region, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2016-01-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Rational improvement of gp41-targeting HIV-1 fusion inhibitors: an innovatively designed Ile-Asp-Leu tail with alternative conformations
Sci Rep, 6, 2016
|
|
5HFL
 
 | |
7MBJ
 
 | |
5WQ2
 
 | |
3PDU
 
 | |
