4AZB
 
 | | Differential inhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH | | Descriptor: | 1,2-ETHANEDIOL, BETA-N-ACETYLHEXOSAMINIDASE, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE, ... | | Authors: | Pluvinage, B, Stubbs, K.A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-06-25 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of the Family 20 Glycoside Hydrolase Catalytic Modules in the Streptococcus Pneumoniae Exo-Beta-D-N-Acetylglucosaminidase, Strh.
Org.Biomol.Chem., 11, 2013
|
|
4AZI
 
 | | Differential inhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH | | Descriptor: | 1,2-ETHANEDIOL, BETA-N-ACETYLHEXOSAMINIDASE, BICINE, ... | | Authors: | Pluvinage, B, Stubbs, K.A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-06-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Inhibition of the Family 20 Glycoside Hydrolase Catalytic Modules in the Streptococcus Pneumoniae Exo-Beta-D-N-Acetylglucosaminidase, Strh.
Org.Biomol.Chem., 11, 2013
|
|
4BQ5
 
 | | Structural analysis of an exo-beta-agarase | | Descriptor: | 3,6-anhydro-alpha-L-galactopyranose, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Pluvinage, B, Hehemann, J.H, Boraston, A.B. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate Recognition and Hydrolysis by a Family 50 Exo-Beta-Agarase Aga50D from the Marine Bacterium Saccharophagus Degradans
J.Biol.Chem., 288, 2013
|
|
4BQ3
 
 | | Structural analysis of an exo-beta-agarase | | Descriptor: | B-AGARASE, CALCIUM ION, GLYCEROL, ... | | Authors: | Pluvinage, B, Hehemann, J.H, Boraston, A.B. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Recognition and Hydrolysis by a Family 50 Exo-Beta-Agarase Aga50D from the Marine Bacterium Saccharophagus Degradans
J.Biol.Chem., 288, 2013
|
|
6ORH
 
 | | Crystal structure of SpGH29 | | Descriptor: | 1,2-ETHANEDIOL, Glycoside hydrolase, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-04-30 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Two complementary alpha-fucosidases fromStreptococcus pneumoniaepromote complete degradation of host-derived carbohydrate antigens.
J.Biol.Chem., 294, 2019
|
|
3LNB
 
 | | Crystal Structure Analysis of Arylamine N-acetyltransferase C from Bacillus anthracis | | Descriptor: | COENZYME A, FORMIC ACID, N-acetyltransferase family protein | | Authors: | Li de la Sierra-Gallay, I, Pluvinage, B, Rodrigues-Lima, F. | | Deposit date: | 2010-02-02 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Bacillus anthracis arylamine N-acetyltransferase ((BACAN)NAT1) that inactivates sulfamethoxazole, reveals unusual structural features compared with the other NAT isoenzymes.
Febs Lett., 585, 2011
|
|
6DRR
 
 | | Crystal structure of Cj0485 dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, Short-chain dehydrogenase | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2018-06-12 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | The gastrointestinal pathogen Campylobacter jejuni metabolizes sugars with potential help from commensal Bacteroides vulgatus.
Commun Biol, 3, 2020
|
|
6DS1
 
 | |
5T7A
 
 | | Crystal structure of Br derivative BhCBM56 | | Descriptor: | 1,2-ETHANEDIOL, BH0236 protein, BROMIDE ION | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-02 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Properties of a family 56 carbohydrate-binding module and its role in the recognition and hydrolysis of beta-1,3-glucan.
J. Biol. Chem., 292, 2017
|
|
6POP
 
 | | Crystal structure of DauA in complex with NADP+ | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase, MAGNESIUM ION, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Insights into the kappa / iota-carrageenan metabolism pathway of some marinePseudoalteromonasspecies.
Commun Biol, 2, 2019
|
|
6PHV
 
 | | SpAga galactose product complex structure | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-galactosidase, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-06-25 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular analysis of an enigmaticStreptococcus pneumoniaevirulence factor: The raffinose-family oligosaccharide utilization system.
J.Biol.Chem., 294, 2019
|
|
6PHY
 
 | | SpAga D472N structure in complex alpha 1,3 galactobiose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-galactosidase, L(+)-TARTARIC ACID, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-06-25 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular analysis of an enigmaticStreptococcus pneumoniaevirulence factor: The raffinose-family oligosaccharide utilization system.
J.Biol.Chem., 294, 2019
|
|
6PI0
 
 | |
6PHW
 
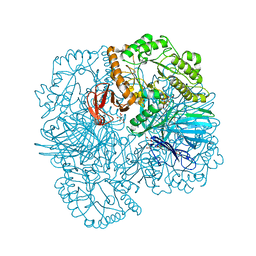 | | SpAga D472N structure in complex with melibiose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-galactosidase, L(+)-TARTARIC ACID, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-06-25 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular analysis of an enigmaticStreptococcus pneumoniaevirulence factor: The raffinose-family oligosaccharide utilization system.
J.Biol.Chem., 294, 2019
|
|
6PNU
 
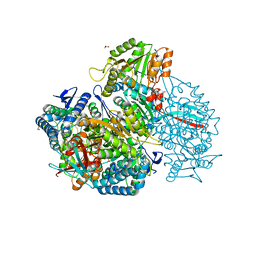 | | Crystal structure of native DauA | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-07-03 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the kappa / iota-carrageenan metabolism pathway of some marinePseudoalteromonasspecies.
Commun Biol, 2, 2019
|
|
6PHU
 
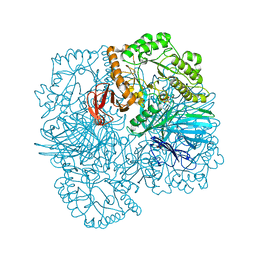 | | SpAga wild type apo structure | | Descriptor: | 1,2-ETHANEDIOL, Alpha-galactosidase, L(+)-TARTARIC ACID | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-06-25 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular analysis of an enigmaticStreptococcus pneumoniaevirulence factor: The raffinose-family oligosaccharide utilization system.
J.Biol.Chem., 294, 2019
|
|
6PHX
 
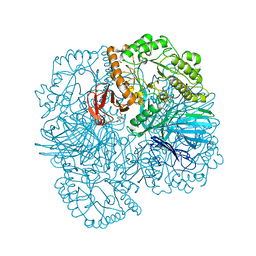 | | SpAga D472N structure in complex with raffinose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Alpha-galactosidase, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-06-25 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular analysis of an enigmaticStreptococcus pneumoniaevirulence factor: The raffinose-family oligosaccharide utilization system.
J.Biol.Chem., 294, 2019
|
|
6ORF
 
 | | Crystal structure of SpGH29 | | Descriptor: | 1,2-ETHANEDIOL, SpGH29, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-04-30 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two complementary alpha-fucosidases fromStreptococcus pneumoniaepromote complete degradation of host-derived carbohydrate antigens.
J.Biol.Chem., 294, 2019
|
|
6ORG
 
 | | Crystal structure of SpGH29 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-04-30 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Two complementary alpha-fucosidases fromStreptococcus pneumoniaepromote complete degradation of host-derived carbohydrate antigens.
J.Biol.Chem., 294, 2019
|
|
6OR4
 
 | | Crystal structure of SpGH29 | | Descriptor: | Glycoside hydrolase, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-04-29 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two complementary alpha-fucosidases fromStreptococcus pneumoniaepromote complete degradation of host-derived carbohydrate antigens.
J.Biol.Chem., 294, 2019
|
|
5FQF
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, FORMIC ACID | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5FQH
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, PHOSPHATE ION | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5FR0
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-deoxy-2-[(difluoroacetyl)amino]-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, PHOSPHATE ION | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-14 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5FQE
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BETA-N-ACETYLGALACTOSAMINIDASE, BROMIDE ION, ... | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5FQG
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, FORMIC ACID | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
