6RQP
 
 | | Steady-state-SMX dark state structure of bacteriorhodopsin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Weinert, T, Skopintsev, P, James, D, Kekilli, D, Furrer, A, Bruenle, S, Mous, S, Nogly, P, Standfuss, J. | | Deposit date: | 2019-05-16 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography.
Science, 365, 2019
|
|
6S0L
 
 | | Structure of the A2A adenosine receptor determined at SwissFEL using native-SAD at 4.57 keV from all available diffraction patterns | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-17 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S19
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 4.57 keV from all available diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.Ch. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S1D
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 4.57 keV from 20,000 diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S1G
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 6.06 keV from 50,000 diffraction patterns. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6RPH
 
 | | TR-SMX open state structure (10-15ms) of bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Weinert, T, Skopintsev, P, James, D, Kekilli, D, Furrer, A, Bruenle, S, Mous, S, Nogly, P, Standfuss, J. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography.
Science, 365, 2019
|
|
6S0Q
 
 | | Structure of the A2A adenosine receptor determined at SwissFEL using native-SAD at 4.57 keV from 50,000 diffraction patterns | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S1E
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 6.06 keV from all available diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6RNJ
 
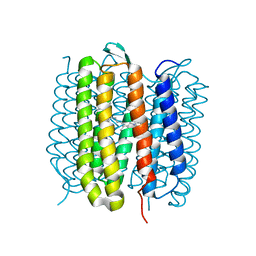 | | TR-SMX closed state structure (0-5ms) of bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Weinert, T, Skopintsev, P, James, D, Kekilli, D, Furrer, F, Bruenle, S, Mous, S, Nogly, P, Standfuss, J. | | Deposit date: | 2019-05-08 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography.
Science, 365, 2019
|
|
6RQO
 
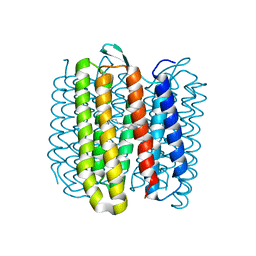 | | Steady-state-SMX activated state structure of bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Weinert, T, Skopintsev, P, James, D, Kekilli, D, Furrer, A, Bruenle, S, Mous, S, Nogly, P, Standfuss, J. | | Deposit date: | 2019-05-16 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography.
Science, 365, 2019
|
|
6QZH
 
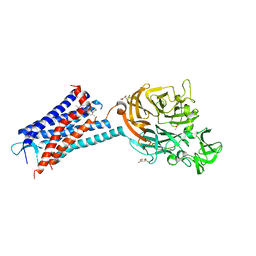 | | Structure of the human CC Chemokine Receptor 7 in complex with the intracellular allosteric antagonist Cmp2105 and the insertion protein Sialidase NanA | | Descriptor: | 3-[[4-[[(1~{R})-2,2-dimethyl-1-(5-methylfuran-2-yl)propyl]amino]-1,1-bis(oxidanylidene)-1,2,5-thiadiazol-3-yl]amino]-~{N},~{N},6-trimethyl-2-oxidanyl-benzamide, C-C chemokine receptor type 7,Sialidase A,C-C chemokine receptor type 7, D(-)-TARTARIC ACID, ... | | Authors: | Jaeger, K, Bruenle, S, Weinert, T, Guba, W, Muehle, J, Miyazaki, T, Weber, M, Furrer, A, Haenggi, N, Tetaz, T, Huang, C.Y, Mattle, D, Vonach, J.M, Gast, A, Kuglstatter, A, Rudolph, M.G, Nogly, P, Benz, J, Dawson, R.J.P, Standfuss, J. | | Deposit date: | 2019-03-11 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Allosteric Ligand Recognition in the Human CC Chemokine Receptor 7.
Cell, 178, 2019
|
|
8QI9
 
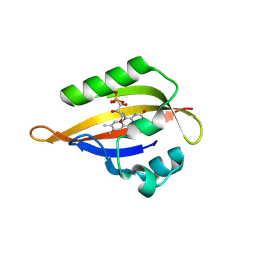 | | CrPhotLOV1 dark state structure determined by serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-11 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
8QIN
 
 | | CrPhotLOV1 light state structure 47.5 ms (45-50 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
8QIP
 
 | | CrPhotLOV1 light state structure 57.5 ms (55-60 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
8QI8
 
 | | Cryogenic temperature dark state structure of CrPhotLOV1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-11 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
8QIH
 
 | | CrPhotLOV1 light state structure 22.5 ms (20-25 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
8QIL
 
 | | CrPhotLOV1 light state structure 37.5 ms (35-40 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
8QIO
 
 | | CrPhotLOV1 light state structure 52.5 ms (50-55 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
8QIR
 
 | | CrPhotLOV1 light state structure 67.5 ms (65-70 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
8QIT
 
 | | CrPhotLOV1 light state structure 77.5 ms (75-80 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
8QIA
 
 | | CrPhotLOV1 light state structure 2.5 ms (0-5 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-11 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
8QIQ
 
 | | CrPhotLOV1 light state structure 62.5 ms (60-65 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
8QIB
 
 | | CrPhotLOV1 light state structure 7.5 ms (5-10 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-11 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
8QIS
 
 | | CrPhotLOV1 light state structure 72.5 ms (70-75 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
8QIK
 
 | | CrPhotLOV1 light state structure 32.5 ms (30-35 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
